Fig. 1.
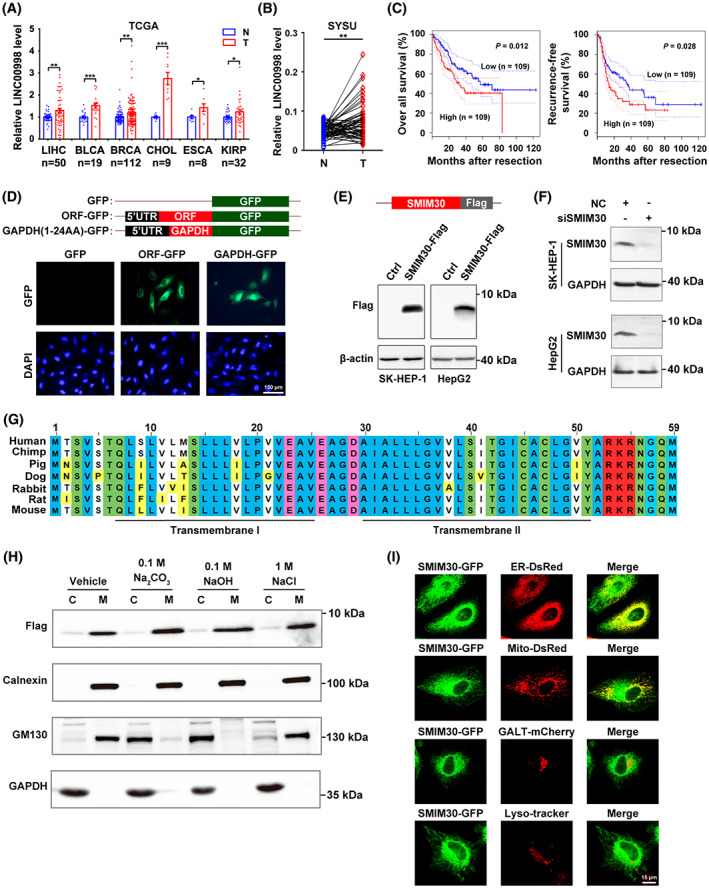
LINC00998‐Encoded transmembrane peptide is located in endoplasmic reticulum and mitochondria and is upregulated in multiple cancer types. (A) The levels of LINC00998 were elevated in multiple cancer types. Pan‐cancer analysis of LINC00998 expression in paired tumor (T) and non‐tumor (N) tissues was performed based on the cancer genome atlas (TCGA) data. LIHC, liver hepatocellular carcinoma (n = 50); BLCA, bladder urothelial carcinoma (n = 19); BRCA, breast invasive carcinoma (n = 112); CHOL, cholangio carcinoma (n = 9); ESCA, esophageal carcinoma (n = 8); KIRP, kidney renal papillary cell carcinoma (n = 32). Data are presented as mean ± SEM, *P < 0.05; **P < 0.01; ***P < 0.001 by two‐sided Student's t‐test. (B) The levels of LINC00998 were increased in HCC tissues. The levels of LINC00998 in 63 paired HCC (T) and adjacent non‐tumor (N) tissues from Sun Yat‐sen University cancer center (SYSU) were assessed by qPCR. **P < 0.01 by two‐sided Student's t‐test. (C) Upregulation of LINC00998 in HCC tissues was associated with worse overall survival and recurrence‐free survival. Analysis was performed using the transcriptome data of human HCC tissues derived from TCGA. Samples with LINC00998 levels in the top 30% (n = 109) and bottom 30% (n = 109) were defined as high‐ and low‐LINC00998 group, respectively. P values were determined by log‐rank test. The dotted lines represent the 95% confidence intervals (CI). (D) The predicted ORF on LINC00998 encoded a micropeptide. Hela cells were transfected with pc3.0‐ORF‐GFP(▵ATG), followed by staining with DAPI and photographed under a fluorescence microscope. pc3.0‐GFP(▵ATG) and pc3.0‐GAPDH‐GFP(▵ATG) were used as a negative and a positive control, respectively. Scale bar, 150 μm. (E) Expression of exogenous flag‐tagged SMIM30. SK‐HEP‐1 or HepG2 cells were transfected with pc3.0‐SMIM30‐flag for 48 h before Western blotting with anti‐flag antibody. β‐Actin, internal control. (F) Expression of cellular endogenous SMIM30 peptide. The indicated cells were transfected with negative control (NC) or siSMIM30 for 48 h before Western blotting with anti‐SMIM30 antibody. GAPDH, internal control. siSMIM30, a mixture of equal amount of siSMIM30‐1 and siSMIM30‐2 which target different regions of SMIM30 mRNA. (G) SMIM30 was highly conserved across mammalian species. Alignment of the predicted amino acid sequences for different mammalian SMIM30 was performed by constraint‐based multiple alignment tool (COBALT) from NCBI website. The amino acids in blue, green, pink, red, and bright yellow indicate the nonpolar, polar, negatively charged, positively charged, and non‐perfectly conserved amino acids, respectively. The two predicted transmembrane helical regions are underlined. (H) SMIM30 was a membrane‐integrated peptide. Homogenates of SK‐HEP‐1 cells that stably expressed flag‐tagged SMIM30 were untreated (vehicle) or incubated with the indicated reagents, and then centrifuged to yield the cytosol (C) or membrane (M) fractions, which were then subjected to immunoblotting analysis. Calnexin, an ER‐localized transmembrane protein, and GM130, a cis‐Golgi tethering protein, were used as the controls for membrane‐integrated molecule and membrane tethering molecule, respectively. (I) SMIM30 was localized in ER and mitochondria. Hela cells were cotransfected with the expression vector of SMIM30‐GFP and the expression construct of either ER‐DsRed, or Mito‐DsRed, or GALT‐mCherry, which marked ER, mitochondria, or Golgi body, respectively. The SMIM30‐GFP‐transfected cells were stained with lysoTracker (red) to mark lysosome. The cells were examined under a confocal microscopy. Scale bar, 15 μm. For D–F and H–I, data are representative of two independent experiments.
