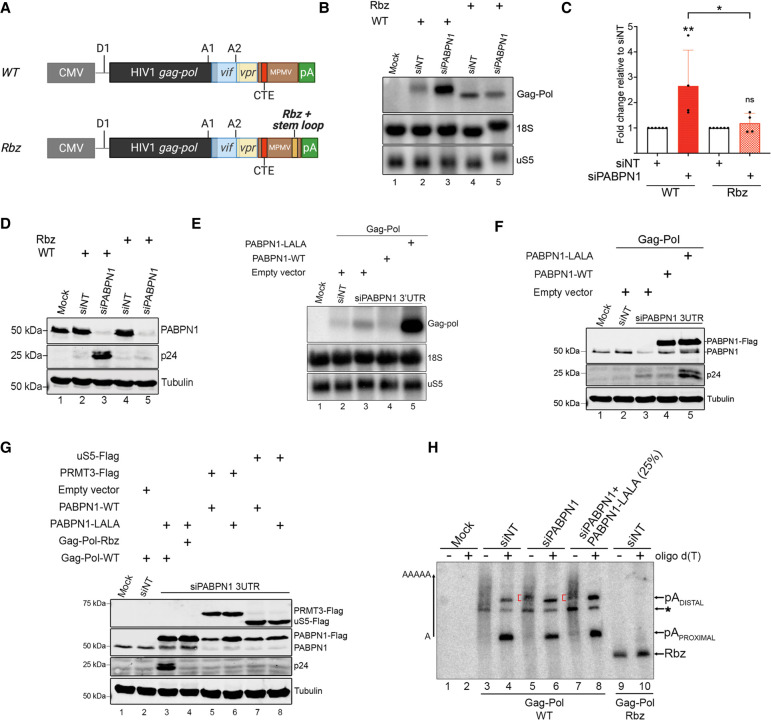
FIGURE 6.
RNA 3′ end processing and polyadenylation is required for PABPN1-dependent repression of unspliced mRNA expression. (A) Schematic of the wild-type (WT) and ribozyme (Rbz) HIV Gag-Pol-CTE reporter contructs showing the insertion of hepatitis delta ribozyme and histone mRNA stabilizing stem–loop (pale orange box) upstream of the MPMV polyadenylation (pA) sites. (B) Northern blot analysis of total RNA prepared from HEK293T cells previously cotransfected with either the wild-type (lanes 2,3) or the ribozyme (lanes 4,5) Gag-Pol-CTE construct and the indicated nontarget (lanes 2,4) and PABPN1-specific (lanes 3,5) siRNAs. The unspliced gag-pol transcript is shown (top). The uS5 mRNA and 18S rRNA were used as loading controls. (C) Levels of unspliced gag-pol transcript were normalized to the levels of uS5 mRNA and expressed relative to values for cells treated with nontarget (siNT) siRNAs. Data and error bars represent the means and standard deviations, respectively, from independent experiments. ns, P-value >0.05; (*) P-value <0.05; (**) P-value <0.01. (D) Western blot analysis of the indicated proteins using extracts prepared from HEK293T cells that were previously cotransfected with either the wild-type (lanes 2,3) or the ribozyme (lanes 4,5) Gag-Pol-CTE construct and the indicated nontarget (lanes 2,4) and PABPN1-specific (lanes 3,5) siRNAs. (E,F) Northern blot (E) and western blot (F) analysis using extracts of HEK293T cells previously cotransfected with the wild-type Gag-Pol-CTE construct (lanes 2–5) and either nontarget (lane 2) or PABPN1-specific (lanes 3–5) siRNAs. Transfections also included constructs expressing either wild-type (lane 4) or LALA (lane 5) versions of PABPN1, as well as an empty vector control (lanes 2,3). (G) Western blot analysis using extracts of HEK293T cells previously cotransfected with nontarget (lane 2) or PABPN1-specific (lanes 3–8) siRNAs and the indicated plasmid constructs. (H) RNase H cleavage assay using total RNA prepared from HEK293T cells transfected with PABPN1-specific (lanes 5–8) and control (lanes 3,4,9,10) siRNAs together with the PABPN1 LALA mutant (lanes 7,8) along with either wild-type (lanes 3–8) or ribozyme (lanes 9,10) Gag-Pol CTE constructs. RNase H assays were performed in the presence (+) and absence (−) of oligo d(T). Due to the significant accumulation of gag-pol transcripts in PABPN1 LALA-expressing cells (part E, lane 5), only 25% of the RNase H assay for the LALA mutant (lanes 7,8) was loaded on gel to compare poly(A) tail distribution with other samples. Red brackets indicate the position of the cluster of PABPN1-dependent 3′ fragments. Transcripts cleaved at the proximal and distal poly(A) sites are indicated on the right. Transcripts cleaved at an unidentified poly(A) site are indicated by an asterisk.