FIGURE 4.
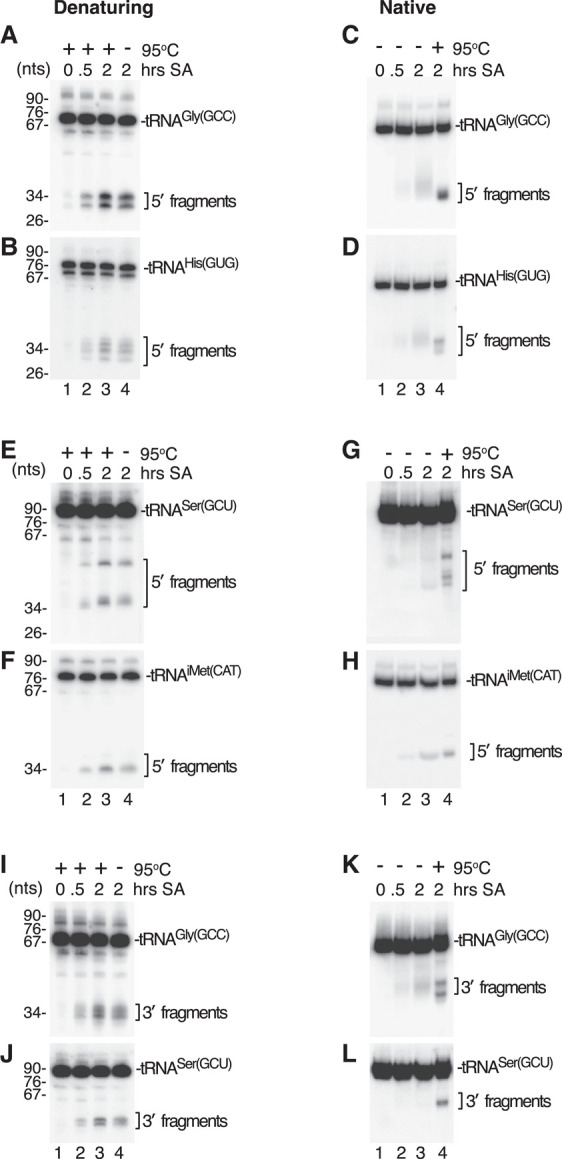
Some human tRNA fragments remain base paired in cells. (A,B) After incubating HEK293A cells with 0.5 M sodium arsenite (SA) for the indicated times, cells were lysed and RNA isolated as described in Materials and Methods and resuspended in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl. After mixing with an equal volume of 95% formamide, 5 mM EDTA and 0.02% bromophenol blue, the RNA was heated to 95°C for 5 min, fractionated in denaturing gels and subjected to northern blotting to detect 5′ halves of tRNAGly(GCC) (A) and tRNAHis(GUG) (B). In lane 4, the heating step was omitted. (C,D) After resuspending in 50 mM Tris-HCl (pH 7.5), 50 mM NaCl, the RNA was mixed with 1/3 vol of 25% glycerol and 0.02% bromophenol blue. After fractionation in native gels, northern blotting was performed to detect 5′ halves of tRNAGly(GCC) (C) and tRNAHis(GUG) (D). In lane 4, the RNA was heated to 95°C for 5 min before loading. (E,F) After heating and fractionation in denaturing gels as in A–B, the indicated RNAs were subjected to northern blotting to detect 5′ halves of tRNASer(GCU) (E) and tRNAiMet(CAT) (F). In lane 4, the heating step was omitted. (G,H) The same RNAs as in E and F were fractionated in native gels followed by northern blotting to detect 5′ halves of tRNASer(GCU) (G) and tRNAiMet(CAT) (H). In lane 4, the RNA was heated to 95°C for 5 min before loading. (I–L) The northern blots in A–B and C–D were reprobed to detect 3′ halves of tRNAGly(GCC) and tRNASer(GCU) after fractionation in denaturing I,J or native gels (K,L).
