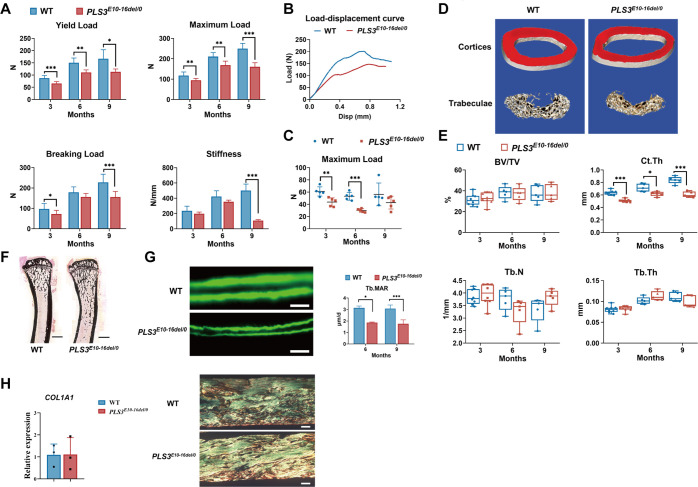
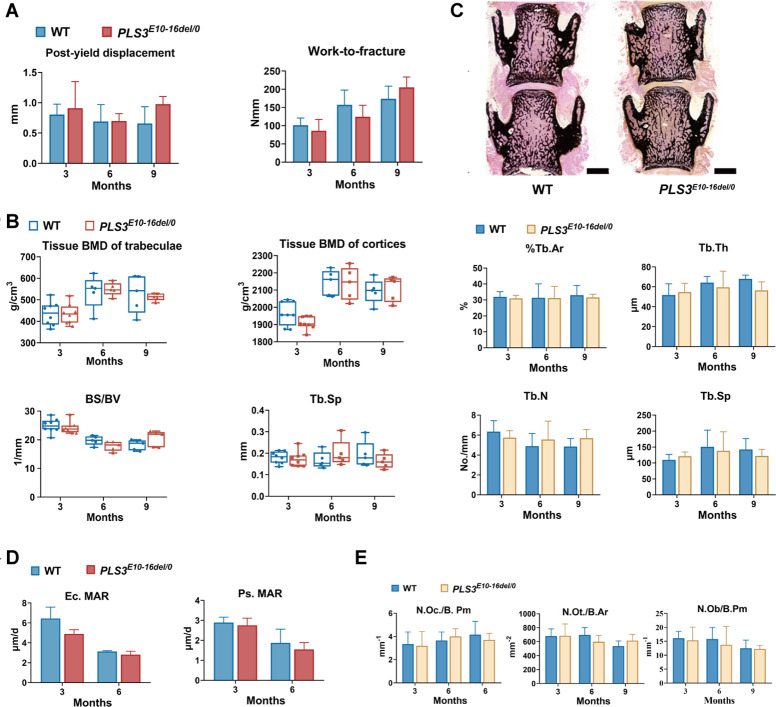
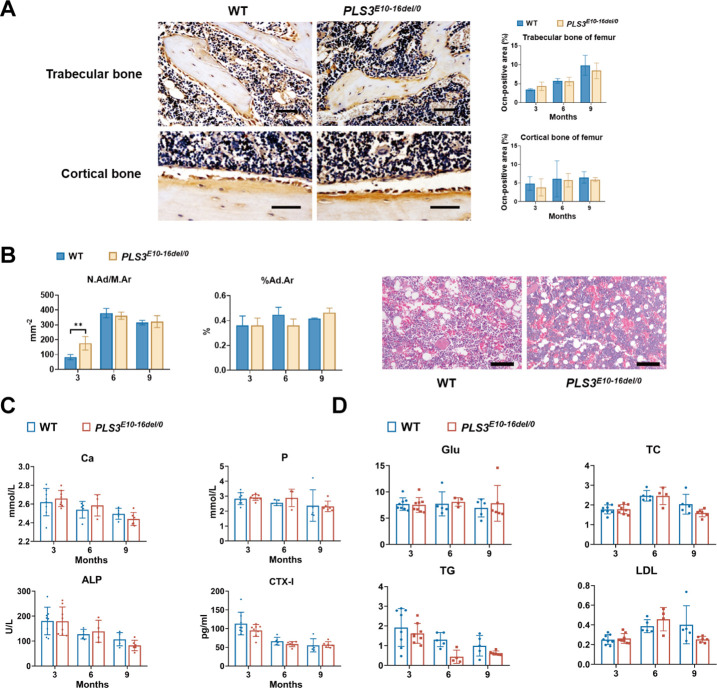
Figure 2. Bone strength, bone microstructure, and bone formation activity of PLS3E10-16del/0 rats.
(A) Mechanical three-point bending tests of femora from PLS3E10-16del/0 and WT rats (n=5-8 per group). (B) Typical load-displacement curves of PLS3E10-16del/0 and WT rats. (C) Indentation tests of L5 from PLS3E10-16del/0 and WT rats (n=5 per group). Data were analyzed using unpaired two-tailed Student t test. *p<0.05, **p <0.01, ***p <0.001 vs WT groups. (D) Three-dimensional reconstruction images of femurs from PLS3E10-16del/0 and WT rats. (E) Micro-CT assessment of the distal femurs from PLS3E10-16del/0 and WT rats (n=5-8 per group). BV/TV: bone volume/tissue volume, Ct.Th: cortical thickness, Tb.Th: trabecular thickness, Tb.N: trabecular number. (F) Representative von Kossa-stained sections of tibia diaphysis of PLS3E10-16del/0 and WT rats. Scale bar = 2000 μm. WT: wild-type. (G) Typical images of unstained and uncalcified vertebra of PLS3E10-16del/0 rats and comparison of mineral apposition rate (n=4 per group). Scale bar = 10 μm. Data were analyzed using unpaired two-tailed Student t test. *p<0.05 vs WT groups. Tb.MAR: mineral apposition rate of lumbar trabeculae. (H) Expression level of COL1A1 and photomicrographs of picrosirius red-stained sections of cortical bone regions of femur visualized through polarized light microscopy. Scale bar = 100 μm. Data were pooled from three independent experiments and were presented as mean ± SEM.