Figure 7.
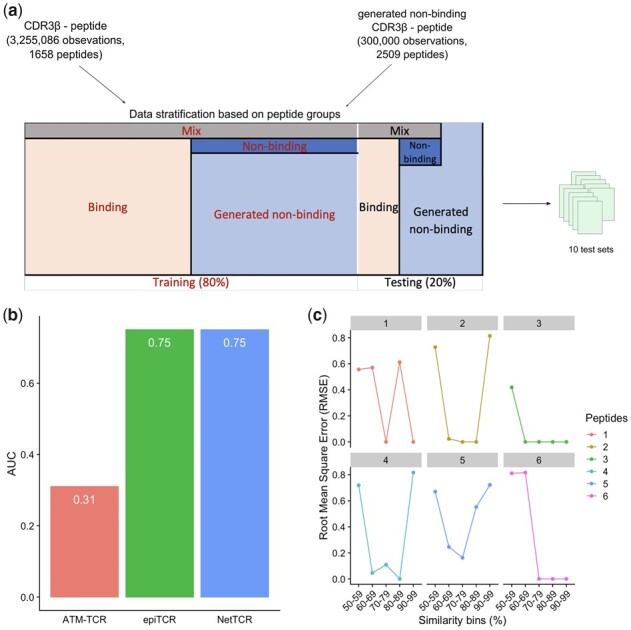
Prediction on observations derived from unseen peptides. (a) Data organization with the use of generated non-binding peptides to balance with the number of binding peptides in training set. (b) The prediction performance (mean AUC) of epiTCR, NetTCR, and ATM–TCR on interactions of unseen peptides from training. (c) Influence of peptide sequence similarity in training set on the predicted labels of peptides in the test set. Six peptides were chosen to represent six groups within the test sets and the proportion of their predicted labels were compared with the proportion of training labels at different levels of Levenshtein similarity using RMSE. The lower the RMSE, the more similar between predicted labels and training labels within a particular bin of peptides at the same level of sequence similarity.
