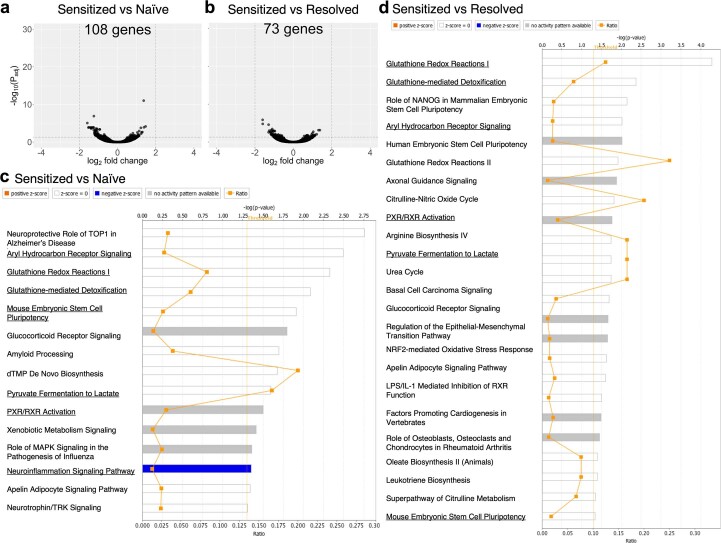
Extended Data Fig. 6. RNA-seq of primary USCs originating from convalescent mice revealed that sensitized USCs maintain differential gene expressions after several passages, related to Fig. 4.
RNAs were isolated from Naïve, Resolved, and Sensitized USCs, then analysed by RNA-seq and performed differential analysis. Significance was determined by Wald test followed by multiple test correction using Benjamini-Hochberg FDR for adjusted p-value.108 and 73 genes were significantly differentially expressed in Sensitized USCs relative to (A) Naïve and (B) Resolved USCs (P-adj < 0.05). Enriched pathways in Sensitized USCs compared with (C) Naïve and (D) Resolved USCs are listed here. Overlapping pathways in both analyses are underlined. IPA determine significance using a right-tailed Fisher’s exact test, with P-adjusted <0.05 considered significantly enriched pathways.