Fig 1. Reconstitution of the ank pathway from A. thermomutatus leads to new natural product.
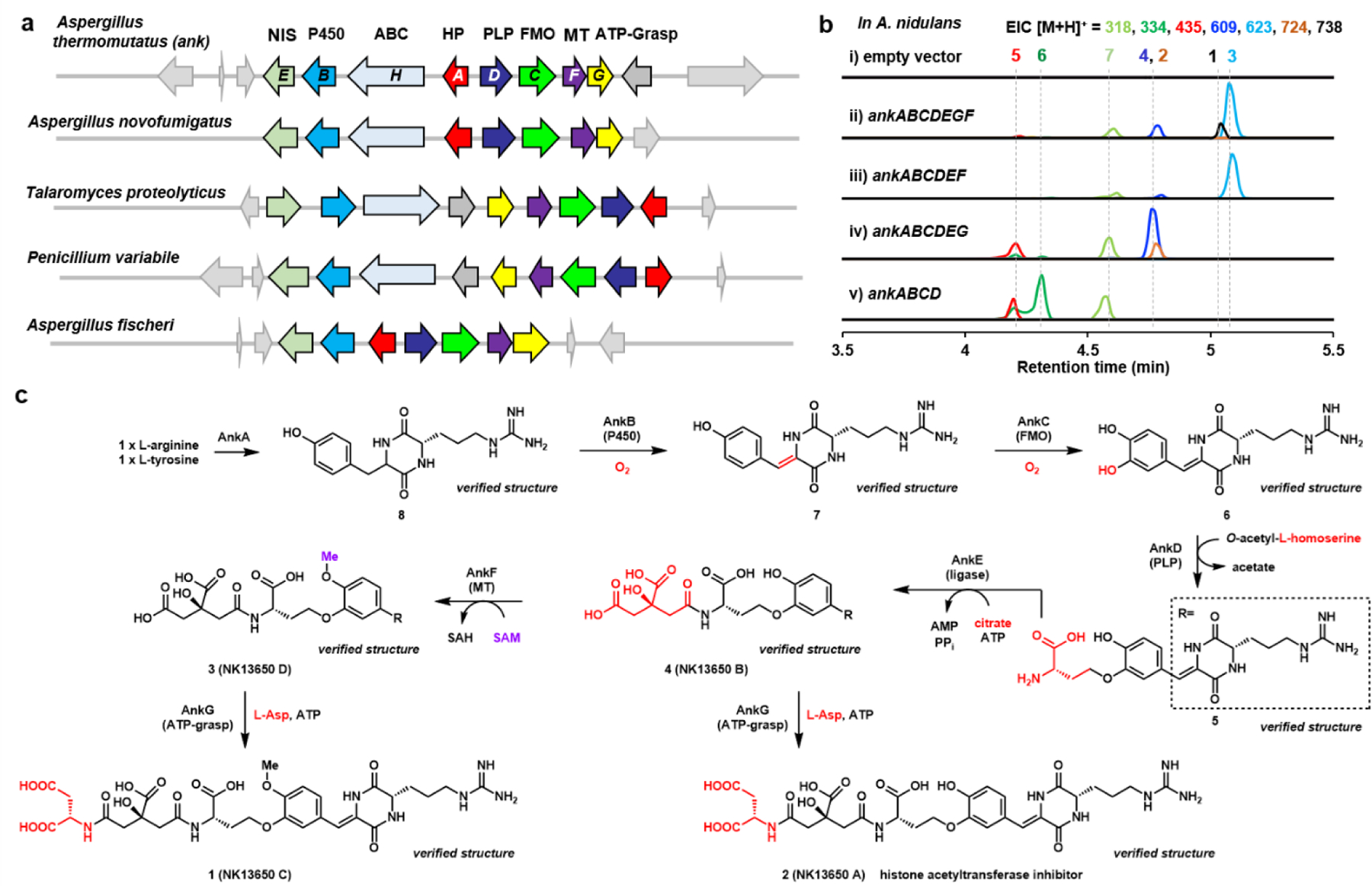
a) The ank BGC encodes the PLP-dependent enzyme AnkD flanked by genes encoding potential tailoring enzymes. This gene cluster is conserved in a number of fungi as shown. Abbreviations: HP: hypothetical protein; P450: cytochrome P450 monooxygenase; FMO: flavin-dependent monooxygnease; PLP: pyridoxal-5’-phosphate dependent enzyme; NIS: NRPS-independent siderophore synthetase. No core enzyme is present, including PKS, NRPS or terpene cyclase. b) QTOF analysis of metabolites produced by different gene combinations in A. nidulans heterologous host are shown in ii) to v). The selected ions are shown and the colors of the traces match to the indicated mass and compounds. A. nidulans transformed with the empty vectors is the control in i). c) Biosynthetic pathway to 1 and 2. The compounds indicated with “verified structure” have been fully characterized by NMR (see Supporting Information). 2 was previously reported to be a p300-selective histone acetyltransferase inhibitor.30
