Fig. 3 |. KDM2A interacts with nucleosome acidic patch.
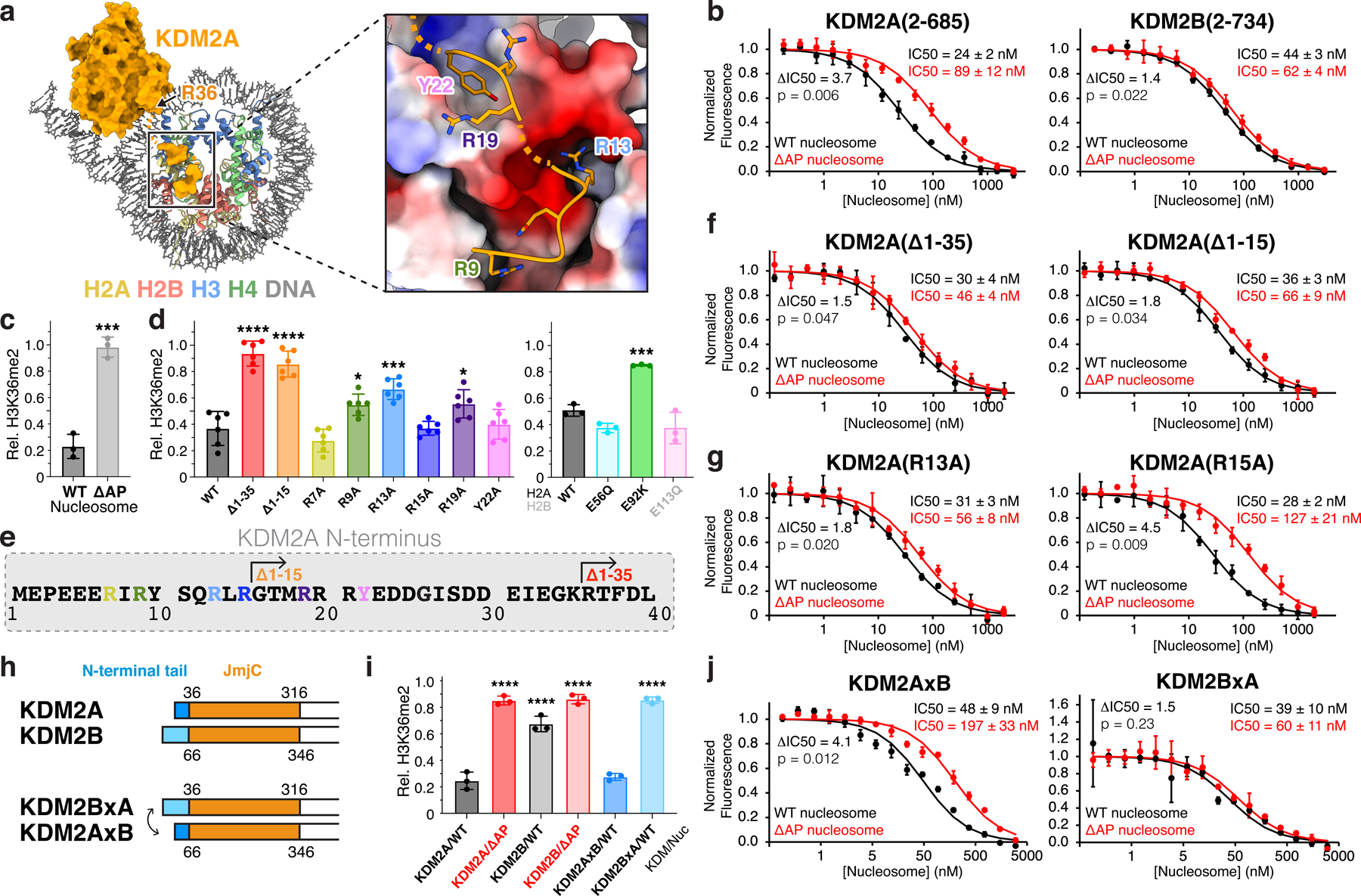
a, N-terminal KDM2A tail residues bind nucleosome acidic patch. b, TR-FRET nucleosome binding competition experiments with KDM2A (left) and KDM2B (right) using wild-type (WT) or acidic patch-neutralized (ΔAP) nucleosomes as competitor. c, KDM2A demethylation of WT and ΔAP H3K36me2-nucleosomes, p = 0.004. d, Demethylation of H3K36me2 nucleosomes by KDM2A N-terminal truncations and mutants (left, p-values for comparison to WT are <0.0001, <0.0001, not significant (ns), 0.468, 0.0002, ns, 0.0361, ns, and <0.0001) and demethylation of oncohistone mutant H3K36me2 nucleosomes by WT KDM2A (right, p-values for comparison to WT are ns, 0.0006, an ns). e, KDM2A N-terminal sequence, with truncations and mutations used in activity and binding assays denoted. f, Nucleosome binding competition experiment with KDM2A truncations using WT or ΔAP nucleosomes as competitor. g, Nucleosome binding competition experiment with KDM2A mutant proteins using WT or ΔAP nucleosomes as competitor. h, Schematic of KDM2 N-terminal tail swap chimeric proteins. i, Demethylation of WT or ΔAP H3K36me2 nucleosomes by WT and chimeric enzymes, KDM2BxA and KDM2AxB, p-values for comparison to KDM2A/WT are <0.0001, <0.0001, <0.0001, ns, <0.0001. j, Nucleosome binding competition experiment with chimeric KDM2 proteins using WT or ΔAP nucleosomes as competitor. For demethylase assays in panels c, d and i, relative residual H3K36me2 (Rel. H3K36me2) plotted as individual data points (n=3 assay technical replicates for panel c and panel d, right, and i, n=6 assay technical replicates for panel d, left) along with mean ± SD, **p ≤0.01, ***p ≤0.001, and ****p ≤0.0001 for statistical differences between WT and mutant samples using an unpaired two-tailed t-test in panel c and an ordinary one-way ANOVA in panels d and i. For binding assays in panels b, f g, and j, mean ± SD are shown (n=3 assay technical replicates) and ratio of ΔAP/WT IC50 (ΔIC50) values and p-values, calculated using an unpaired two-tailed t-test, indicated.
