Fig. 4 |. Dynamic and multivalent KDM2A-nucleosome interactions.
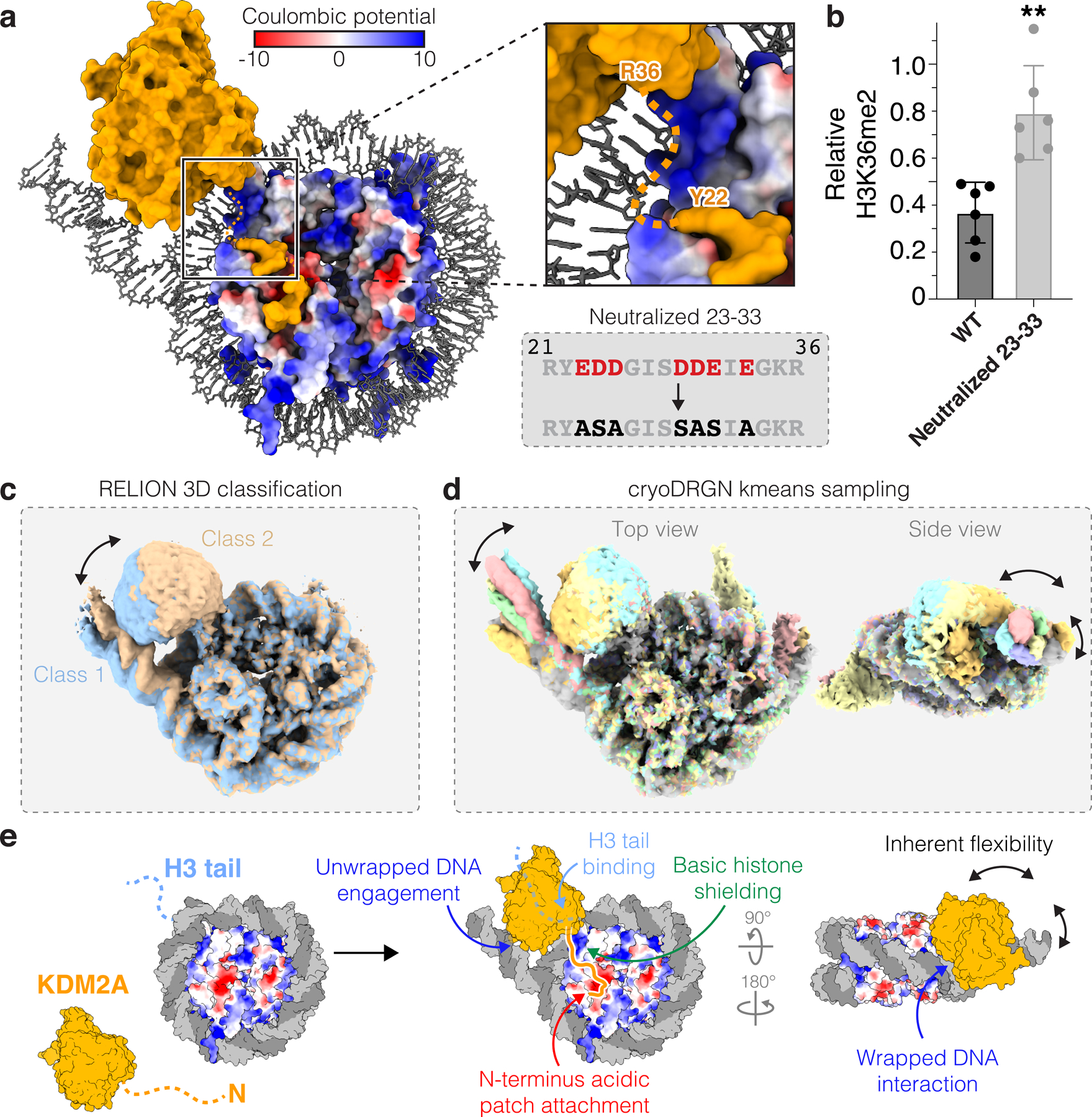
a, Basic histone surface exposed by DNA unwrapping between JmjC domain and acidic patch-binding region of KDM2A. b, Demethylation of H3K36me2-nucleosomes by WT and neutralized 23–33 mutant KDM2A. Relative residual H3K36me2 plotted as individual data points (n=6 assay technical replicates) along with mean ± SD, **p ≤0.01 (0.0014) using an unpaired two-tailed t-test. c-d, 3D classification in RELION (c) and heterogeneity analysis with cryoDRGN kmeans sampling (d) reveals correlated heterogeneity in regions of structure corresponding to JmjC domain and unwrapped DNA. e, Model of KDM2A-nucleosome binding includes dynamic and multivalent interactions with multiple nucleosome surfaces.
