Figure 1. Elevated levels of VLCFAs induce S1P production and neuronal dysfunction.
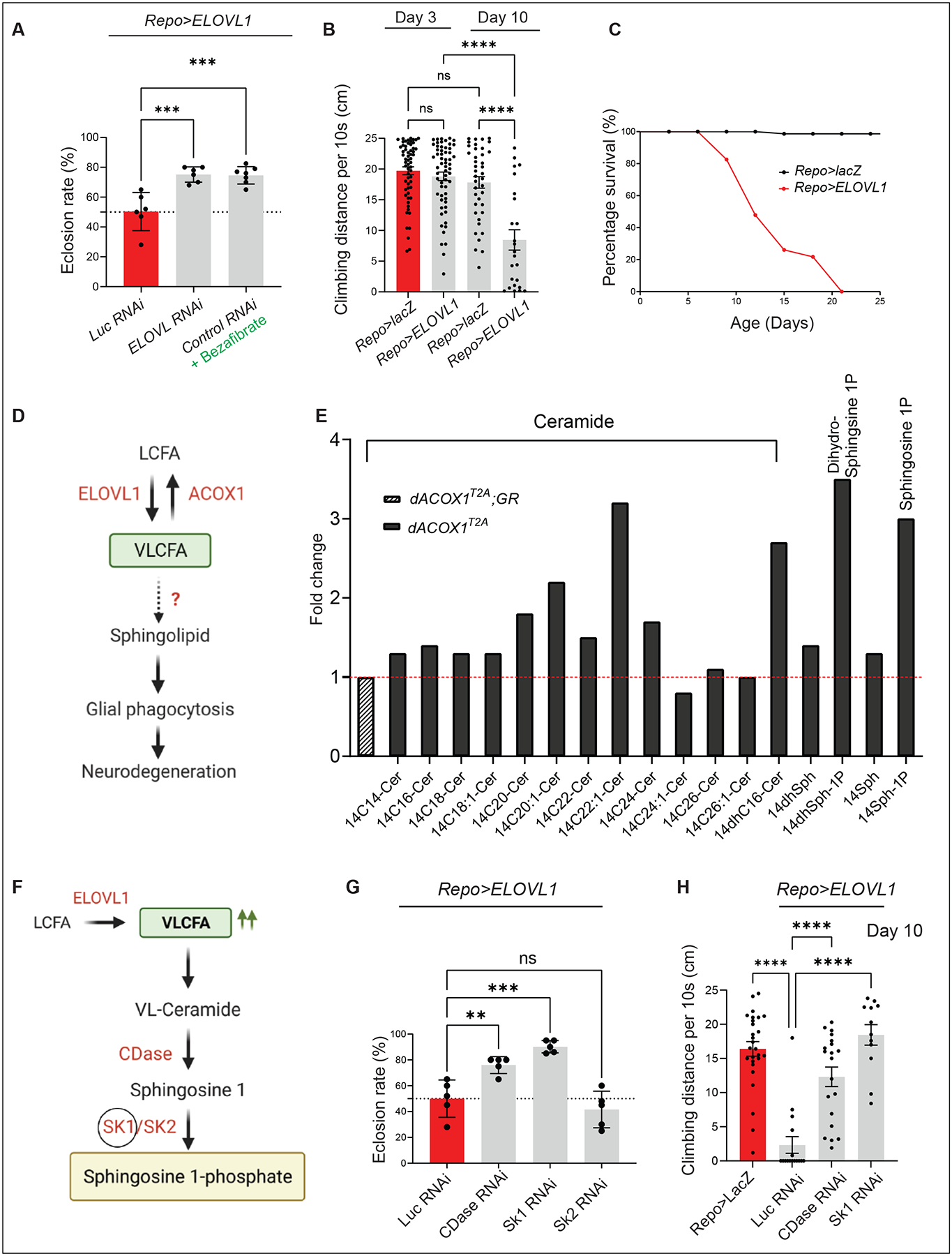
(A) Expressing ELOVL RNAi to downregulate the fly homologue of ELOVL1 or Bezafibrate supplementation suppresses the low eclosion rate observed in Repo>ELOVL1 flies. Quantification of the percentage of expected animals per cross (n>6). (B) Glial ELOVL1 expression causes progressive climbing defects (n>24) and (C) significantly decreases lifespan (n=100 for Repo>lacZ and Repo>ELOVL1). (D) Model of the mechanisms of VLCFA leading to neurodegeneration. (E) Sphingolipid profiling in heads of dACOX1T2A mutants (n=500 per each genotype). Cer: Ceramide, Sph: Sphingsoine, dhSph: dihydro Sphingosine, Sph-1P: Sphingosine 1-phosphate, dhSph-1P: dehydro sphingosine 1-phosphate. GR: Genomic Rescue construct (F) Pathway to convert VLCFAs into S1P. (G) A decrease in the levels of CDase or SK1 but not SK2 significantly suppresses the lethality observed in Repo>ELOVL1 flies. Quantification of the percentage of expected animals per cross (n=5 per each genotype) (H) A decrease in the levels of CDase or SK1 significantly suppresses the progressive climbing defects observed in Repo>ELOVL1 flies (n>11). Statistical analyses are one-way ANOVA followed by a Tukey post hoc test. Results are mean ± s.e.m. (****p < 0.0001, ***p < 0.001, **p < 0.01; n.s., not significant).
