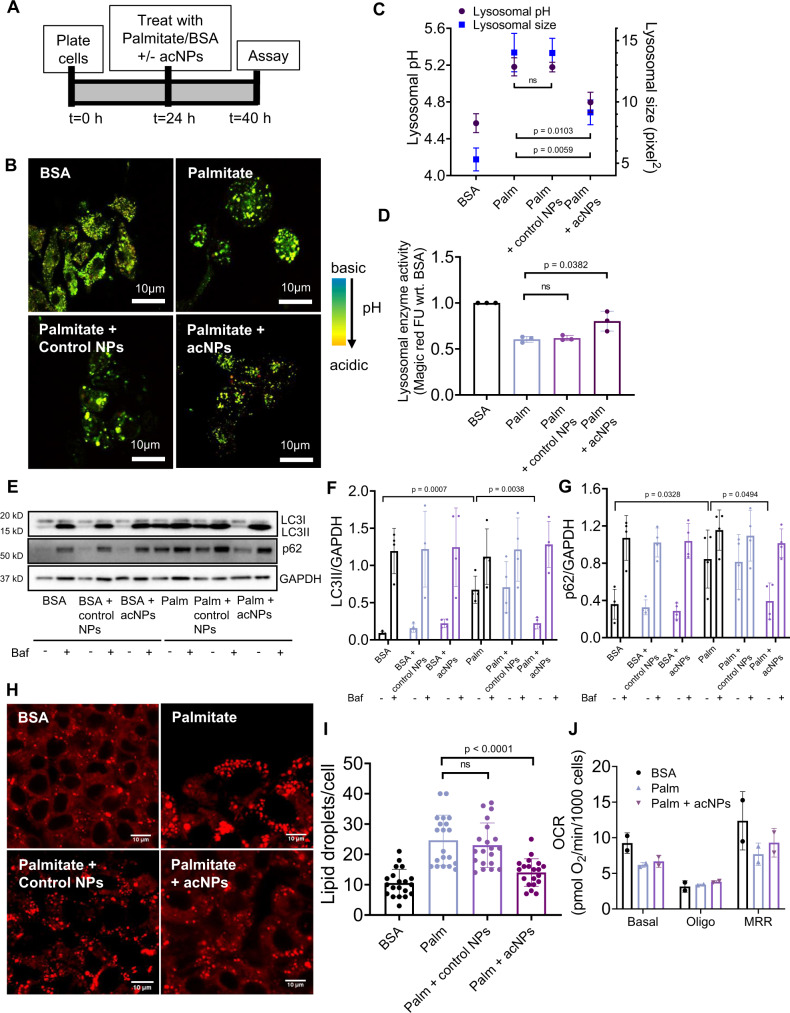
Fig. 3. Treatment of HepG2 with acNPs reverses the defects in lysosomal acidity, cathepsin L activity and autophagic flux induced by lipotoxicity.
A Schematic of experimental protocol for cell treatment for 16 h before assaying for lysosomal acidity, autophagy, or cellular function. The indicated conditions are control BSA, 400 µM palmitate complexed to BSA, or 400 µM palmitate with control NPs and acNPs treatment. The 16 h timepoint is chosen because it shows the highest amount of autophagic flux inhibition in HepG2 cells under palmitate. B Representative confocal microscopy images of HepG2 cells treated with the indicated conditions and stained with 75 nM pH-sensitive Lysosensor dye to assess lysosome acidity. Bar, 10 μm. C Mean lysosomal pH and lysosomal area per cell for cells exposed to the indicated conditions were analyzed by MetaMorph® show significant restoration of lysosomal pH compared with palmitate cells (N = 3 independent experiments with n = 20 cells analyzed per condition). D Assessment of lysosomal cathepsin L activity by Magic red fluorescent substrate assay in HepG2 cells exposed to all the conditions showed significant restoration of lysosomal enzyme activity with acNPs treatment but not with control NPs treatment (N = 3 independent experiments). E–G Representative western blot exposure images showing protein expression data for control NPs and acNPs treated to HepG2 cells, before and after addition of Bafilomycin A1 (Baf) (N = 4 independent experiments). H Representative confocal images of HepG2 cells stained with Nile Red dye for 15 min and imaged with fluorescence microscopy. Nile red dye accumulated quickly in the lipid vesicles. Bar, 10 μm. I Quantification of lipid vesicles number indicated significant reduction in lipid droplets density after acNPs treatment in HepG2 cells exposed to palmitate. Control NPs addition did not reduce lipid droplets (n = 20 cells analyzed per condition). J Mitochondria oxygen consumption rates in HepG2 cells under BSA, palmitate or palmitate with acNPs (N = 3 independent experiments). Two-tailed unpaired t-test (C, D, F, G, I, J); data are expressed as means ± SD, n.s. not statistically significant. Source data are available as a Source Data file.