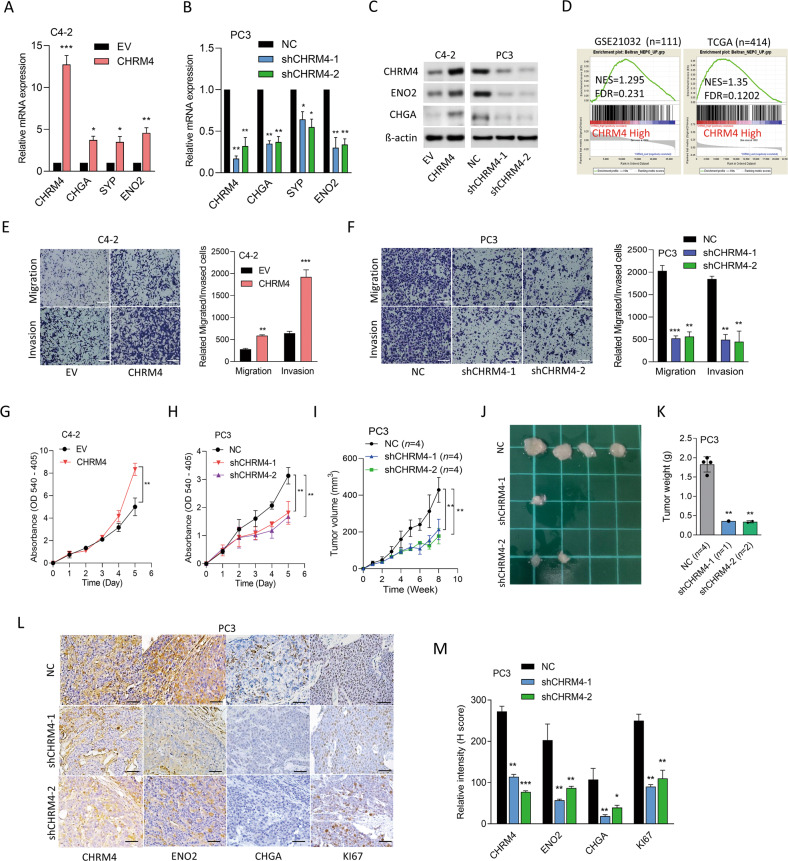
Fig. 3. Increased CHRM4 is associated with oncogenic features and neuroendocrine differentiation in prostate cancer.
A, B CHRM4 and NE marker (CHGA, SYP, and ENO2) mRNA levels in C4-2 cells stably transfected with an empty vector (EV) or a CHRM4-expressing vector (A) or in PC3 cells stably transfected with the non-targeting control (NC) or CHRM4 shRNA vector (B), measured by an RT-qPCR analysis, * vs. the EV (A) or NC (B), by a one-way ANOVA. C Western blot showing CHRM4 and NE marker protein levels in CHRM4-modified C4-2 and PC3 cells. D GSEA analysis of TCGA prostate cancer dataset revealed positive correlations between higher CHRM4 expression in prostate tissues and gene profiles reflecting NEPC-responsiveness. NES, normalized enrichment score; FDR, false discovery rate. E–H Relative cell migration and invasion through Matrigel (E, F) and proliferation (G, H) of CHRM4-overexpressing C4-2 (E, G) or CHRM4-knockdown (KD) PC3 cells (F, H). n = 5 per group. * vs. the EV (E, G) or NC (F, H), by a one-way ANOVA. I–K Tumor growth analysis (I, J) and tumor weights (K) of CHRM4-KD PC3 cells subcutaneously inoculated into male nude mice for 8 weeks. Tumor weights were measured on the day tumors were collected. Tumor sizes were measured once a week and analyzed by a one-way ANOVA. IHC staining (L) and representative intensities (M) of CHRM4, ENO2, CHGA, and KI67 in subcutaneous tumors from J. * vs. NC-bearing tumors, by a two-tailed Student’s t-test. Quantification of relative mRNA levels, and migration, invasion, and proliferation are presented as the mean ± SEM from three biological replicates. *p < 0.05, **p < 0.01, ***p < 0.001.