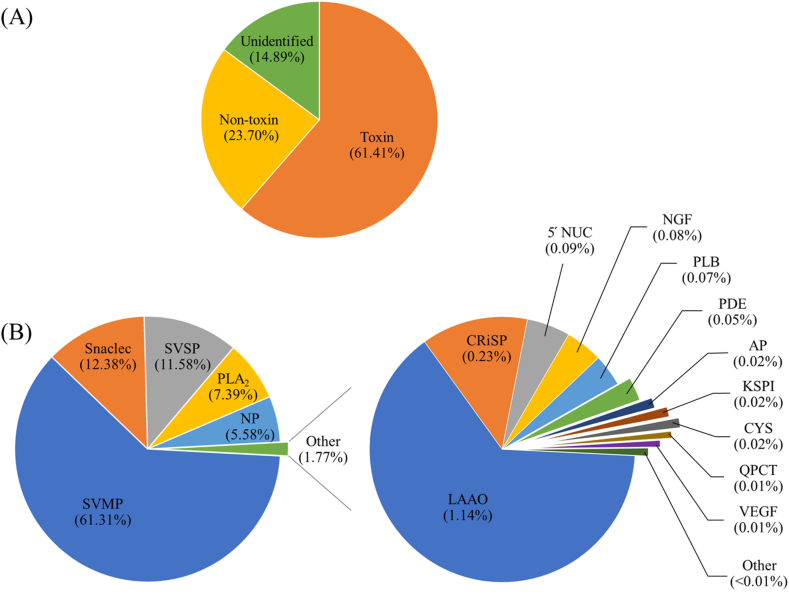
Fig. 1.
De novo transcriptome of C. rhodostoma venom glands. (A) The pie chart displaying an overview of transcript-cleaned reads with FPKM ≥1 identified by the NCBI non-redundant (nr) protein, Swiss-Prot, and Pfam databases. (B) Pie charts representing the expression abundance (%) of toxin transcripts by gene families. SVMP, snake venom metalloproteinase, demonstrates the most abundance, followed by Snaclec, snake venom C-type lectin/lectin-like protein; SVSP, snake venom serine protease; PLA2, phospholipase A2; and NP, natriuretic peptide, respectively (B, left). The other transcripts from the toxin families were minor (1.77%), and consist of LAAO, L-amino acid oxidase; CRiSP, cysteine-rich secretory protein; NUC, 5′ nucleotidase; NGF, nerve growth factor; PLB; phospholipase B, PDE, phosphodiesterase; AP, aminopeptidase; KSPI, Kunitz-type serine protease inhibitor; CYS, cystatin; QPCT; glutaminyl-peptide cyclotransferase, VEGF, vascular endothelial growth factor. While HYA, hyaluronidase; PLA2I, phospholipase A2 inhibitor; DPP IV, dipeptidylpeptidase IV; CVF, Cobra venom factor; WAP, waprin; and AChE, acetylcholinesterase, were expressed at levels lower than 0.01% of FPKM from identified toxins (B, right).