Figure 1.
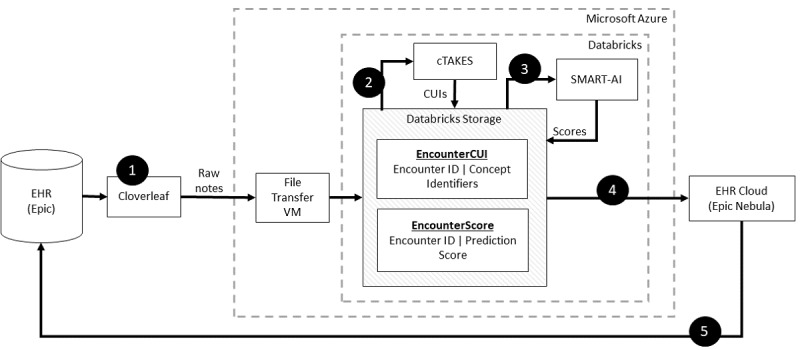
Step-by-step implementation of clinical natural language processing (NLP) pipeline. Step 1 ran a scheduled program to ingest notes from the EHR for each patient organized the notes, and relayed them via an HL7 data feed (Cloverleaf) into the cloud computing environment and data lake (Microsoft Azure and Databricks) onto a VM (Step 2). The NLP engine (cTAKES) processed the text stored on the VM and mapped them to medical concepts from the National Library of Medicine’s metathesaurus (CUIs). The machine learning model received the CUIs as inputs and stored the results in DataBricks. At regular intervals, a custom Python script in Databricks performed the text extraction and linguistic feature engineering via cTAKES and stored CUIs with the appended data of patient identifiers. The CUIs served as the input to the machine learning model (SMART-AI) at the encounter level. The output of prediction probabilities and classification was stored in a Databricks table (Step 3). An API call from the EHR cloud is made to determine whether the cutpoint threshold from the machine learning model was met to trigger a BPA. In Step 4, the EHR cloud made an HTTP call to Databricks to request the score. The score was returned to the EHR cloud and subsequently delivered as a BPA when the provider opened the patient’s chart in our on-premise instance of the EHR (Step 5). API: application programming interface; BPA: best practice alert; cTAKES: Clinical Text Analysis and Knowledge Extraction System; CUI: concept unique identifier; EHR electronic health record; HL7: Health Level 7; SMART-AI: Substance Misuse Algorithm for Referral to Treatment Using Artificial Intelligence; VM: virtual machine.
