Fig. 3.
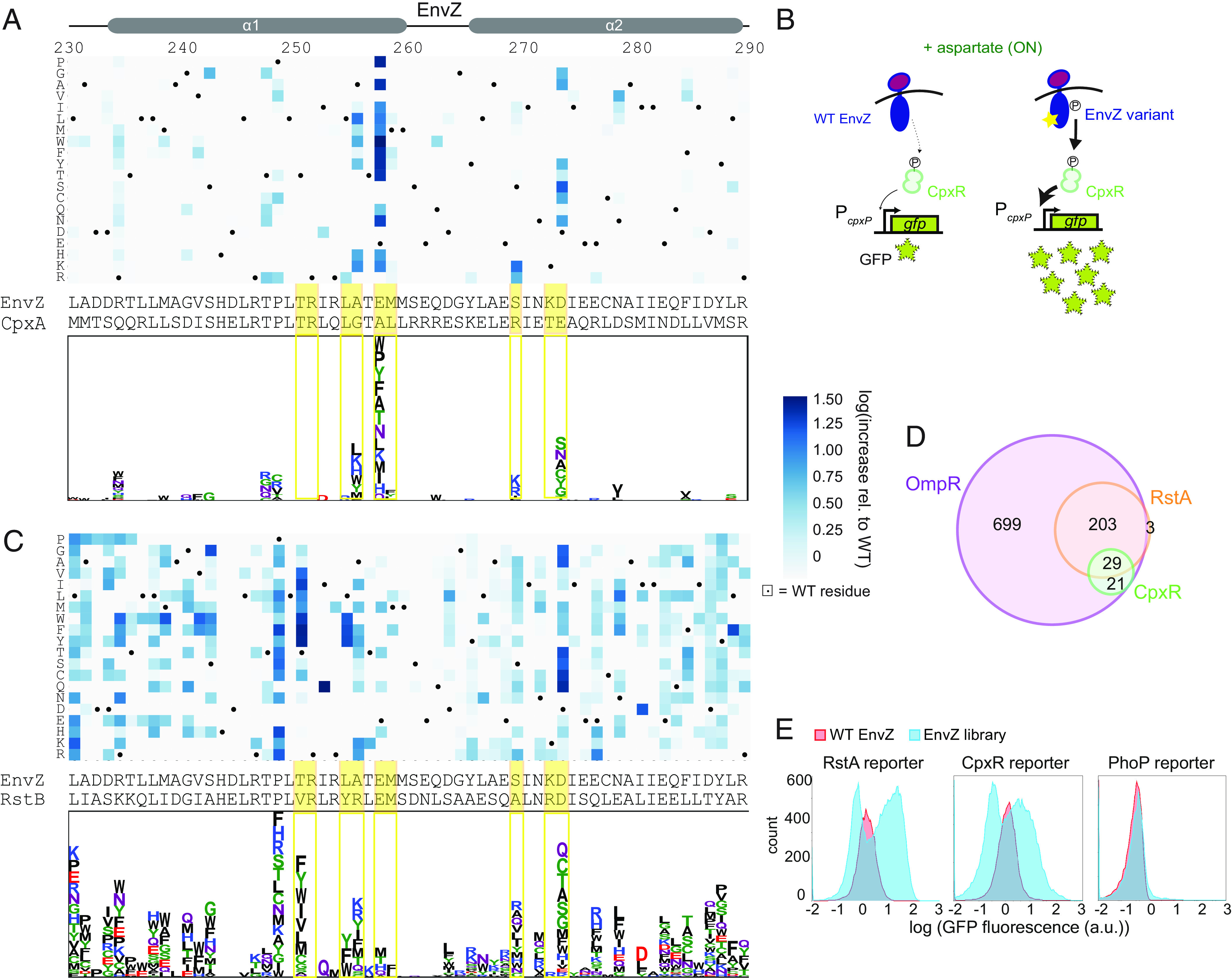
EnvZ exhibits marginal specificity, reflecting a crowded local sequence space. (A) Top: Heatmap of CpxR reporter data where values represent increases in GFP fluorescence relative to wild-type EnvZ in the +signal condition. Dots mark wild-type residues. EnvZ and CpxA DHp sequences are shown below. Yellow highlighted positions mark coevolving residues. Values were normalized by increases in fluorescence toward OmpR, which may reflect nonspecific effects of a substitution on expression level or kinase activity that increase activity toward all RRs. Bottom: Logo transformation of heatmap where the height of a letter represents the increase in fluorescence relative to wild-type for that amino acid substitution at that position. Letters are stacked in ranked order. (B) Diagram illustrating fluorescence measurements for CpxR reporter. Wild-type EnvZ shows low levels of kinase activity, leading to low GFP levels. EnvZ variants (as indicated by the star) may show increased kinase activity, driving high GFP production. (C) Same as (A) except for RstA reporter data. (D) Overlap of single-substitution EnvZ variants with activity toward different RRs. The OmpR set contains variants with kinase activity for OmpR comparable to wild-type EnvZ (within 5-fold). The RstA and CpxR sets contain variants with ≥5-fold increases in activity toward RstA or CpxR relative to wild-type EnvZ. (E) Histograms of GFP fluorescence distributions for the RstA, CpxR, and PhoP reporters. Red populations are for wild-type EnvZ, blue populations are transformations of the single mutant library. Blue library populations for RstA and CpxR reporters are replicated from Fig. 1C for comparison to PhoP.
