Fig. 5.
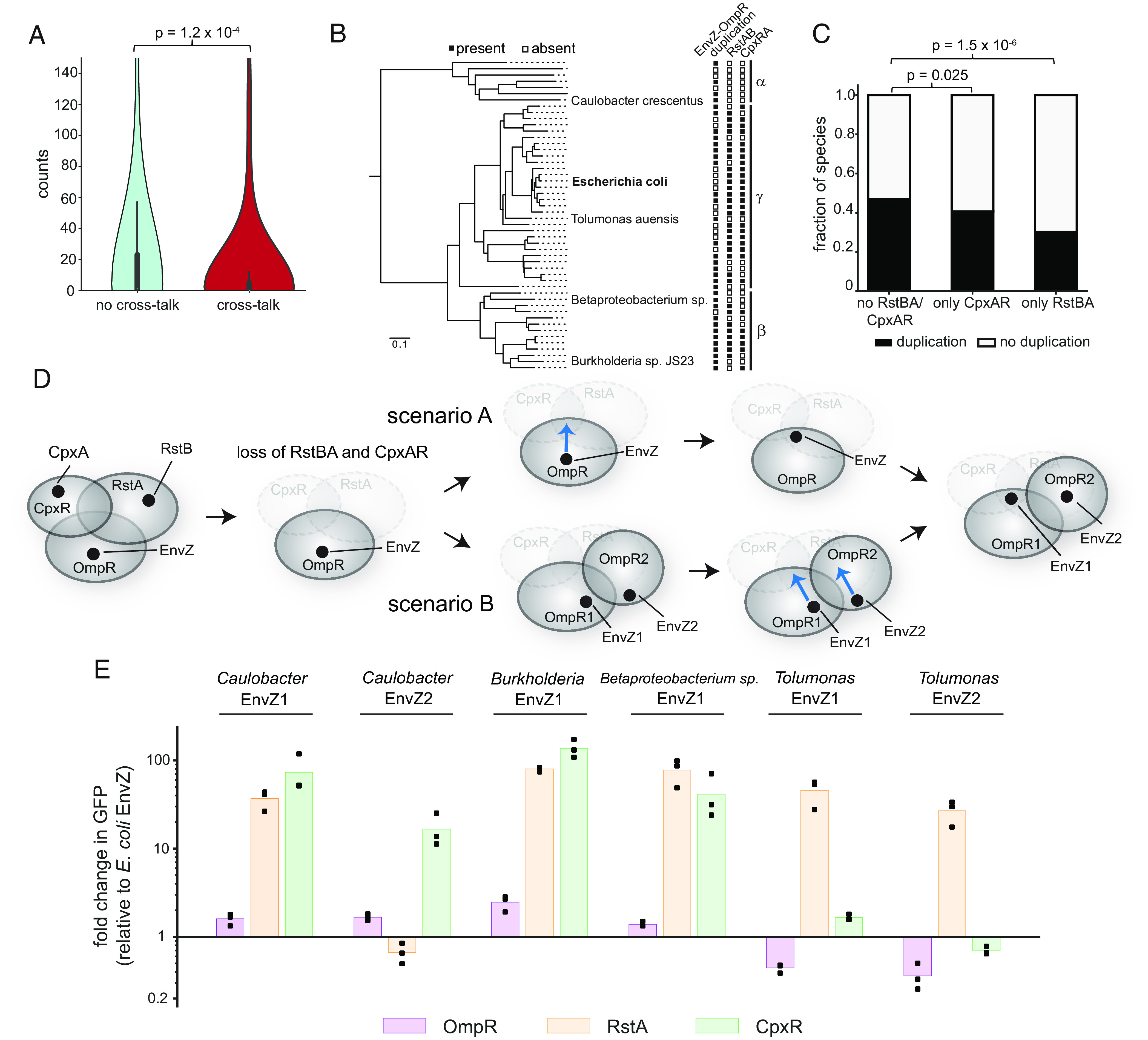
A densely occupied local sequence space constrains the evolution of two-component systems. (A) Violin plots show the distributions of counts of single substitutions found at the equivalent position in 1,019 EnvZ orthologs from species that also have RstBA and CpxAR. Counts are shown for two categories of substitution: those which do produce cross-talk to either RstA or CpxR and those which do not (see Fig. 3, P = 1.2 × 10−4, Kolmogorov–Smirnov test). The inner box shows the quartiles and the whiskers show the range except for outliers. (B) Tree of a subset of species indicating whether they have RstBA and CpxAR and whether they have EnvZ–OmpR duplications. Closed and open squares indicate presence and absence, respectively. (C) Fraction of species that either do not have RstBA or CpxAR, have only CpxAR, or have only RstBA, that have duplications of EnvZ–OmpR (P = 0.025 for CpxAR loss, P = 1.5 × 10−6 for RstBA loss, Fisher’s exact test). (D) Sequence space diagram illustrating how species that have lost RstBA and CpxAR may relax the selection pressure against entering regions of sequence space that would previously have resulted in cross-talk, also freeing up sequence space for EnvZ–OmpR duplications. Blue arrow indicates a single mutation in EnvZ that would be tolerated only in species that have lost CpxR and RstA. This could occur before duplication (scenario A) or after (scenario B). (E) Fold changes in fluorescence relative to E. coli EnvZ for six EnvZ homologs from four distantly related species, for E. coli OmpR, RstA, and CpxR reporters. n = 3 biological replicates.
