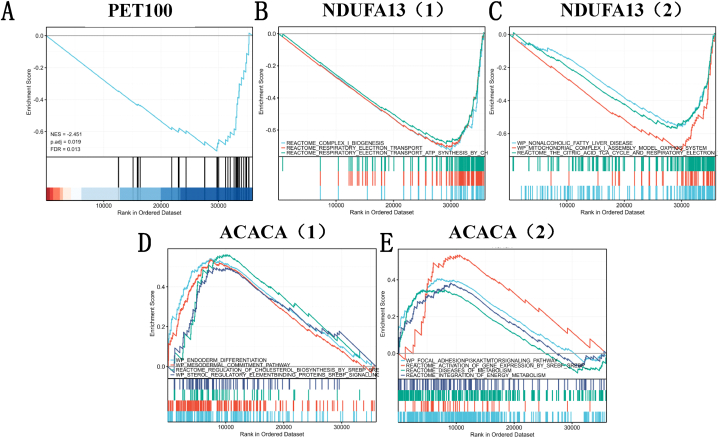
Fig. 11.
EP300 GSEA is used to annotate the malignant hallmarks of these genes with different expression levels. (A) PET100; (B)–(C) NDUFA13; (D)–(E) ACACA. The abscissa is the sorted gene list, and the ordinate is the value of signal2 noise. The area ratio of gray shadow can reflect the size of signal2 noise between groups as a whole, that is, the difference between. (Here, it is considered that the threshold of significant enrichment is: false discovery rate (FDR) < 0.25 and p. adjust <0.05).