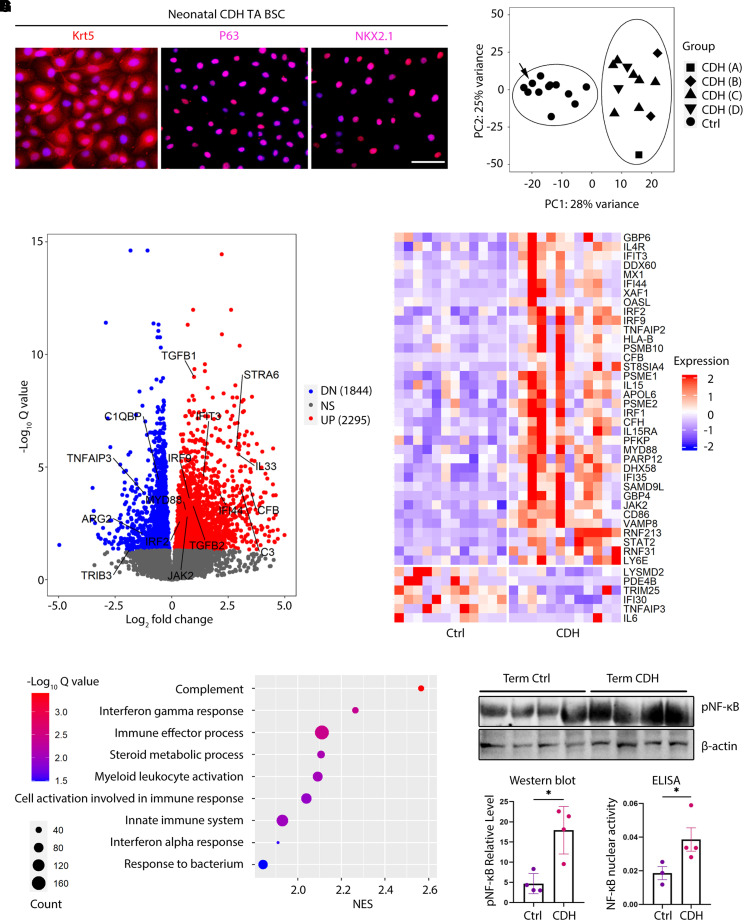
Figure 2.
Congenital diaphragmatic hernia (CDH) basal stem cells (BSCs) have a disease-associated transcriptome and exhibit a proinflammatory signature. (A) Representative images of antibody staining for KRT5, TP63, and NKX2.1 in BSCs derived from TA of newborns with CDH. (B) Principal component analysis of bulk RNA sequencing datasets of BSC samples from CDH newborns (n = 12) and control, term (n = 6), and preterm (n = 6) newborns. Patients with CDH are labeled with A, B, C, and D according to their reported type of defect size during surgical repair. Arrow indicates one control case with prenatal oligohydramnios-associated lung hypoplasia. (C) Volcano plot highlighting differentially expressed genes between CDH and control BSCs with an adjusted P < 0.05. (D) Pathway analysis for bulk RNA sequencing results of CDH and control BSCs. (E) Heatmap of interferon signaling pathway genes that are differentially expressed between CDH and control BSCs. (F) Representative Western blot for concentrations of Ser536 phosphorylation of the p65 subunit of nuclear factor kappa B (pNF-ΚB) in CDH (n = 4) and control (n = 4) BSCs. β-actin was a loading control. (G) Activity of nuclear NF-kB measured by ELISA in CDH BSCs (n = 4) and control BSCs (n = 3). *P < 0.05 by Mann-Whitney U test. Scale bar, 25 μm. Ctrl = control; DN = downregulated; NS = not significant; pNF-ΚB = phosphorylated nuclear factor kappa B; TA = tracheal aspirate; UP = upregulated.