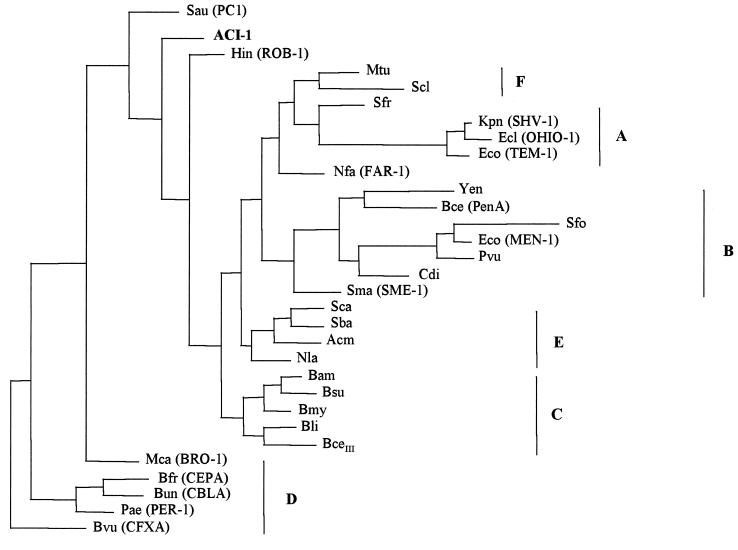
FIG. 4.
Phylogenetic trees obtained for 31 class A β-lactamases, according to the parsimony criteria. Abbreviations: Sau, S. aureus PC1 (P00807); Hin, H. influenzae ROB-1 (P33949); Mtu, M. tuberculosis (Q10670); Scl, S. clavuligerus (Z54190); Nfa, N. farcinica FAR-1 (AF024601); Acm, Actinomadura sp. strain R39 (X53650); Sca, S. cacaoi (P14560); Sba, S. badius (P35391); Nla, N. lactamdurans (Q06316); Bam, B. amyloliquefaciens (Q44674); Bsu, B. subtilis (P39824); Bmy, B. mycoides (P28018); Bli, B. licheniformis (P00808); BceIII, B. cereus β-lactamase type III (P06548); Yen, Y. enterocolitica (Q01166); Bce, B. cepacia (U85041); Sfo, S. fonticola (P80545); Eco (MEN-1), E. coli (P28585); Cdi, C. diversus (P22390); Pvu, P. vulgaris (P52664); Sma, S. marcescens Sme-1 (P52682); Sfr, S. fradiae (P35392); Kpn, K. pneumoniae SHV-1 (P23982); Ecl, E. cloacae (OHIO-1) (P18251); Eco (TEM-1), E. coli (P00810); Mca, M. catarrhalis (BRO-1) (Q59514); Bvu, B. vulgatus (CfxA) (P30899); Bfr, B. fragilis (CepA) (L13472); Bun, B. uniformis (CblA) (P30898); Pae, P. aeruginosa (PER-1) (P37321). The codes in parentheses correspond to listed accession numbers (see reference 23 and the European Bioinformatics Institute website [http://www.ebi.ac.uk]).