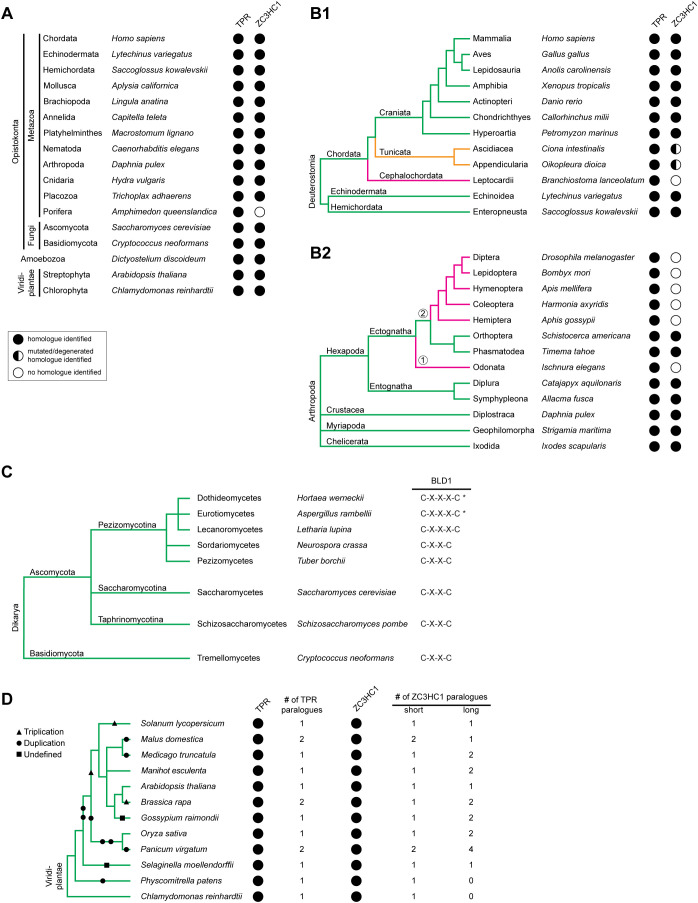
FIGURE 3:
Distribution of ZC3HC1 and its homologues among eukaryotes. (A) Selection of eukaryotic phyla and divisions with representative species in which ZC3HC1 homologues were identified by sequence database mining. The presence of TPR homologues is depicted for comparison. Taxonomy is mostly according to the NCBI taxonomy database (Schoch et al., 2020; https://www.ncbi.nlm.nih.gov/taxonomy). Note that ZC3HC1 homologues exist in most animal phyla and the divisions of fungi, amoeba, and green plants. Among the listed animal phyla, ZC3HC1 was undetectable only in the sponges. (B) Exemplifying cladograms of the clade Deuterostomia and the phylum Arthropoda, illustrating the evolutionary fate of ZC3HC1 in some subphyla. (B1) Cladogram of the Deuterostomia, based on phylogenetic trees and former cladograms (e.g., Delsuc et al., 2018), listing the phyla Echinodermata, Hemichordata, and Chordata, the chordate subphyla Craniata, Tunicata, and Cephalochordata, and a selection of classes and species. Note that ZC3HC1 homologues were undetectable in the Cephalochordata, while homologues in the Tunicata harbor various mutations that differ between different classes and orders, but would all prevent HsZC3HC1 from binding to the NB and TPR. (B2) Cladogram of the phylum Arthropoda, deduced from a phylogenetic tree (Sasaki et al., 2013), listing the subphyla Hexapoda, Crustacea, Myriapoda, and Chelicerata, and a selection of orders and species. Note that ZC3HC1 homologues were undetectable in the infraclass Paleoptera (①), here represented by the order Odonata, and are similarly absent in most orders of the infraclass Neoptera (②). However, ZC3HC1 homologues are present in the orders Orthoptera and Phasmatodea, suggesting that at least two independent events led to the disappearance of the ZC3HC1 gene, or its alteration beyond recognition, in the other insects. (C) Cladogram of the subkingdom Dikarya, representing an excerpt of a previous cladogram (Spatafora et al., 2017), with its divisions Basidiomycota and Ascomycota. For the Ascomycota, three subdivisions are listed, i.e. the Pezizomycotina, Saccharomycotina, and Taphrinomycotina, together with some of their classes and representative species. While most fungal ZC3HC1 homologues possess the C-X(2)-C tetrapeptide as part of their BLD1 zinc finger, some Pezizomycotina classes feature a C-X(3)-C instead. However, even though the C-X(3)-C pentapeptide is predominant in the classes Eurotiomycetes and Dothideomycetes, a few of their orders feature only sequences with C-X(2)-C. These classes are therefore marked with an asterisk. (D) Cladogram of the kingdom Viridiplantae, representing an excerpt of a phylogenetic tree (Panchy et al., 2016; Wang et al., 2020), including some green plant orders in which genome duplication events did or did not occur. Representative species and genome duplication (circles), triplication (triangles), and undefined polyploidization events (squares) are indicated. Several genome duplications led to six ZC3HC1 and two TPR paralogues in the order Poales, here represented by the switchgrass Panicum virgatum. By contrast, in other orders, the additional ZC3HC1 gene appears to have been inactivated again, for example, in the spreading earth moss Physcomitrella patens.