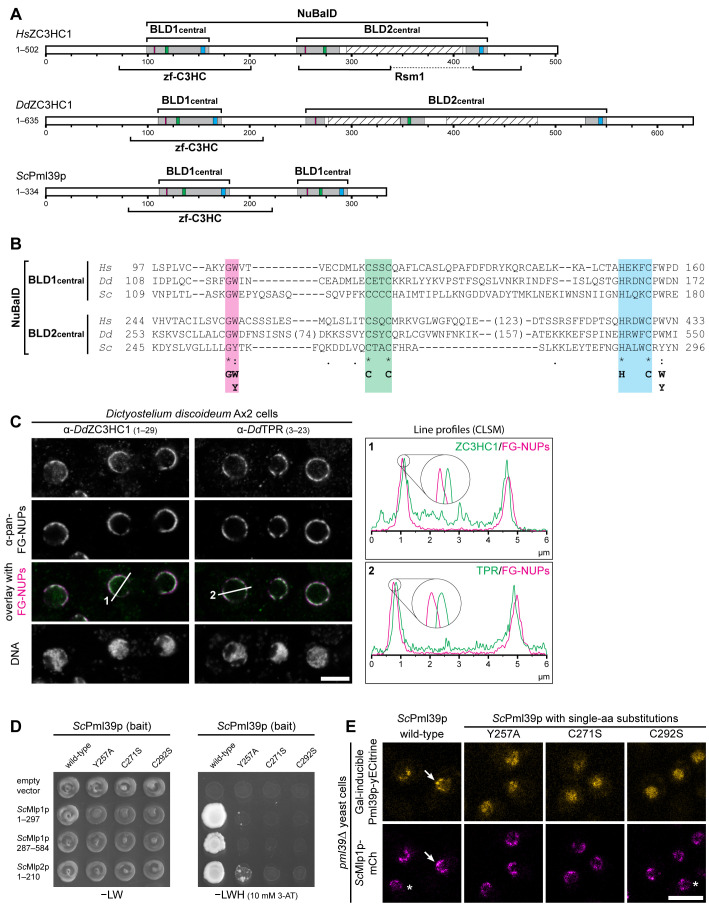
FIGURE 4:
NB- and TPR/Mlp-interacting ZC3HC1 homologues with a conserved NuBaID signature exist in S. cerevisiae and D. discoideum. (A) Schematic depiction of DdZC3HC1 and ScPml39p, compared to HsZC3HC1. The highlighted regions of the two BLDs, together representing the bimodular NuBaID, correspond to those in Figures 1C and 2A1. The boxes in magenta now depict positions that can read either G-W or G-Y, the latter dipeptide part of the Pml39p BLD2. The known NLS of HsZC3HC1 and the unknown, only conjecturable NLS of the two other homologues appear differently positioned and are not depicted here. The expanse of the minimal central region of each HsZC3HC1 BLD is as specified in Figure 2A2. The central regions of DdZC3HC1 BLD1 and BLD2 shown here comprise I108–N172 and K253–I550, respectively. The inner boundaries flanking two apparent insertions within DdZC3HC1 BLD2 here correspond to S273 and D348, and to K371 and A529. Parts of these insertions were predicted to be mostly unstructured, except for V418–N433, and to range from N277 to K347 and N393 to S482 (hatched boxes). For ScPml39p, the central BLD1 and BLD2 regions presented here comprise V109–E180 and K245–N296, respectively. Brackets indicate regions to which a zf-C3HC or Rsm1 motif has been attributed to date. So far, no Rsm1 motif has been assigned to DdZC3HC1 and ScPml39p. (B) Sequence alignments of the central regions of the BLDs, according to those in Figure 2A2. The minimal sequence signature shared by the two BLDs in all three proteins is depicted. (C) Double-labeling IFM of D. discoideum Ax2 cells with pan-FG-NUPs antibodies and antibodies for either DdZC3HC1 or DdTPR, with the focal plane approximately at the equator of most nuclei. DNA staining is shown for reference. Section lines across the nuclei, marked 1 and 2 in the overlay micrographs, were analyzed by ImageJ, with line profiles plotted. Note that the 4× enlarged line profiles from both sides of the corresponding nuclei reveal the offset location of DdZC3HC1 and DdTPR toward the nuclear interior, relative to the immunolabeled FG-repeat nucleoporins of the NPCs. Bar, 5 µm. (D) Representative Y2H data, obtained by expressing segments of Mlp1p and Mlp2p with either intact Pml39p or a selection of Pml39p mutants with single-aa substitutions in the NuBaID. Experiments were performed as described in Figure 2C. Note that while a robust Y2H interaction occurred between intact Pml39p and distinct parts of the Mlps, no colony growth was observed for Pml39p mutants such as Y257A, C271S, and C292S. (E) Live-cell imaging of pml39∆ yeast cells endogenously expressing mCherry-tagged Mlp1p and, upon induced ectopic expression, either yECitrine-tagged WT Pml39p or a selection of the likewise tagged mutants with single-aa substitutions. Note that the newly synthesized intact Pml39p primarily accumulated at the NE (arrow). By contrast, the Pml39p mutants were distributed throughout the nuclear interior. Asterisks mark a few cells in which Pml39p expression was not detected. Bar, 5 µm.