FIGURE 6:
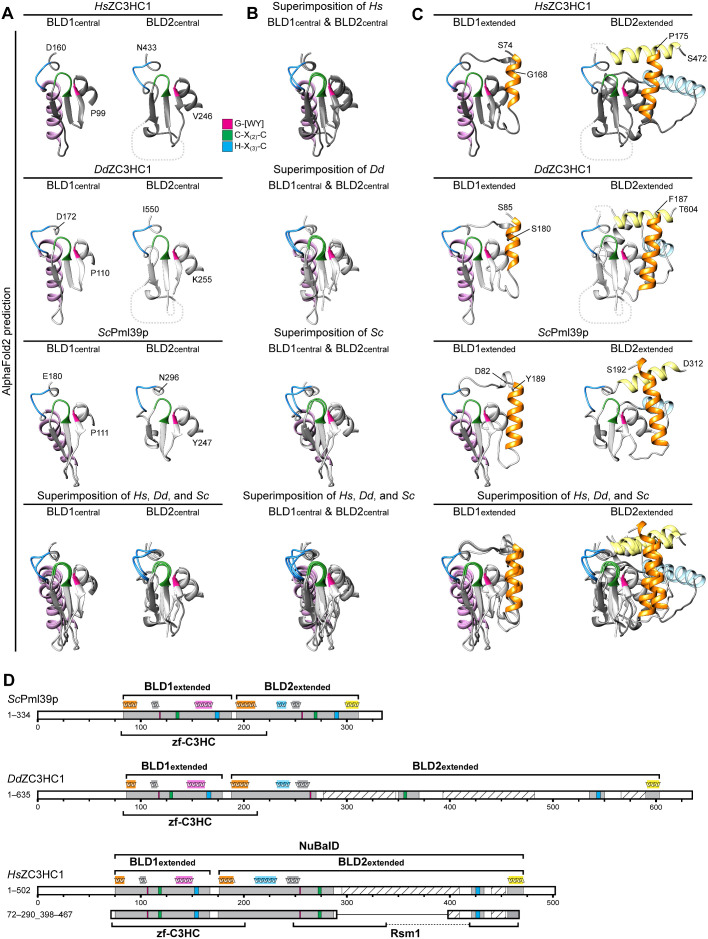
Tertiary structure predictions by AlphaFold2 uncover striking similarities between the BLDs of HsZC3HC1, DdZC3HC1, and ScPml39p and allow the redefinition of their boundaries. (A) Structures predicted for the central regions of the BLD1 and BLD2 of HsZC3HC1, DdZC3HC1, and ScPml39p. The outer boundaries of the central part of BLD1 shown here are P99 and D160 for HsZC3HC1, P110 and D172 for DdZC3HC1, and P111 and E180 for ScPml39p. The outer boundaries of the central part of BLD2 here correspond to V246 and N433 for HsZC3HC1, K255 and I550 for DdZC3HC1, and Y247 and N296 for ScPml39p. Having blinded out the AlphaFold2 prediction for the major loop-like BLD2 insertion of HsZC3HC1 and the two major insertions of DdZC3HC1 BLD2, the inner BLD2 boundaries shown here correspond to I287 and F418 for HsZC3HC1, and to I270 and K350, and I370 and E535, respectively, for DdZC3HC1. However, the relative positions of the blinded-out loops are depicted as dashed lines (not to scale). The HsZC3HC1 and ScPml39p structures, here and in B and C, were obtained from the AlphaFold database. The structure for the sequence-corrected version of DdZC3HC1 (accession number ON368701) was determined using the AlphaFold2 source code. The sequence elements C-X(2)-C and H-X(3)-C, assumed to be involved in zinc ion coordination, and the G-[WY] dipeptide are colored as in Figure 4A. Note the similarities between the central BLD1 structures of the homologues and those between the central BLD2 structures. Also note the BLD1-specific α-helix, colored in light pink. (B) Superimposition of the central parts of BLD1 and BLD2 onto each other. Aside from the evolutionarily conserved BLD1-specific α-helix, the structural similarity between the other central parts of both BLDs, which are considered involved in zinc ion coordination, appears evident. (C) Structural predictions for essentially the entire BLD1 and BLD2 modules as newly defined in our study. An additional residue was appended to each boundary to facilitate recognition. The BLD1 of HsZC3HC1, comprising K75–F167, is thus presented as S74–G168. The HsZC3HC1 BLD2, comprising A176–S471, is shown as P175–S472, yet with the major loop and now also a smaller second loop between I434 and E455 blinded out as in A. Accordingly, the BLD1 of DdZC3HC1 is presented as S85–S180, instead of N86–F179, and its BLD2 as F187–T604, instead of Q188–S603. Again, the two major loops within the amoebic BLD2 have been blinded out, as was a smaller loop between V551 and I589. BLD1 and BLD2 of ScPml39p, defined as L83–E188 and S193–E311, are presented correspondingly as D82–Y189 and S192–D312. The BLD1-specific α-helix is again colored in light pink, while the α-helices specific for the BLD2 of all three homologues are shown in light yellow and light blue. The α-helix common to the N-terminal boundary of both BLDs is highlighted in orange. As an aside, note that all segments shown here primarily comprise residues for which AlphaFold2 assigned, with only a few exceptions, a high per-residue confidence score of at least 70, mostly exceeding 90. (D) Schematic depiction of HsZC3HC1, DdZC3HC1, and ScPml39p with the newly defined BLD boundaries. These schemes depict an additional minor insertion within the BLD2 of HsZC3HC1 (I434–E455) and DdZC3HC1 (V551–I589), with the predicted unstructured regions (S440–A454 of HsZC3HC1, G566–S590 of DdZC3HC1) again shown as hatched. For simplification, other potentially unstructured regions beyond the outer BLD boundaries and found in all three homologues are not highlighted. The schematic indications of the α-helices above the scheme for each homologue represent the relative positions of the α-helices correspondingly colored in C. According to the novel BLD delineations, the zf-C3HC motif of the Pfam database would now comprise sequences encompassing the entire BLD1 and part of BLD2. The Rsm1 motif, assigned to HsZC3HC1 and not to DdZC3HC1 or ScPml39p, corresponds only to parts of the BLD2 and its loop-like insertions. Finally, note that the minimal NE binding–competent HsZC3HC1 mutant 72–290_398–467, schematically depicted here for comparison, comprises, with the exception of the four residues 468–471, the newly defined BLD regions in their entirety.