Abstract
Horizontal transfer of transposable elements (TEs) is an important mechanism contributing to genetic diversity and innovation. Bats (order Chiroptera) have repeatedly been shown to experience horizontal transfer of TEs at what appears to be a high rate compared with other mammals. We investigated the occurrence of horizontally transferred (HT) DNA transposons involving bats. We found over 200 putative HT elements within bats; 16 transposons were shared across distantly related mammalian clades, and 2 other elements were shared with a fish and two lizard species. Our results indicate that bats are a hotspot for horizontal transfer of DNA transposons. These events broadly coincide with the diversification of several bat clades, supporting the hypothesis that DNA transposon invasions have contributed to genetic diversification of bats.
Keywords: endogenous retrovirus, fusogenic envelope protein, monotremes, echidna, platypus
Introduction
Transposable elements (TEs), DNA fragments that can mobilize within and across genomes, comprise most horizontally transferred (HT) genetic material in eukaryotes (Wallau et al. 2012; El Baidouri et al. 2014). Although viruses are prime candidates as TE vectors (Gilbert et al. 2010, 2014, 2016; Thomas et al. 2010), the exact mechanisms of how TEs are transferred and invade the germline of eukaryotes are unclear. Nevertheless, horizontal transfer of TEs (HTT) into naïve genomes can allow TEs to successfully invade and propagate before the host can effectively silence the invaders with anti-TE defenses (Schaack et al. 2010; Kofler et al. 2018). Class II elements (DNA transposons and rolling-circle [RC] elements), particularly Tc-Mariner transposons, are overrepresented in eukaryote HT events compared with Class I elements (retrotransposons) (Peccoud et al. 2017; Zhang et al. 2020), likely due to differences in mobilization mechanisms allowing easier transmission (Lampe et al. 1996; Silva et al. 2004; Gilbert et al. 2016; Gilbert and Feschotte 2018; Palazzo et al. 2019).
The activity and repetitive nature of TEs have shaped genome structure and phenotypes in diverse lineages, by increasing TE copy number, introducing genetic diversity, altering regulatory networks, promoting shuffling of exons, and introducing TE domains that can be co-opted by the host genome (Feschotte and Pritham 2007; Feschotte 2008; Cordaux and Batzer 2009; Schaack et al. 2010; Casacuberta and González 2013; Thomas et al. 2014; Grabundzija et al. 2016; Zhang et al. 2019; Cosby et al. 2021). Yet the magnitude of influence on genome evolution in mammals is unclear, as previous studies were limited by relatively few mammal genome assemblies and TE data sets. High sequence similarity among observed DNA transposons and relatively recent divergence of many mammal lineages make it difficult to parse HTT versus vertical inheritance (Gilbert et al. 2010; Novick et al. 2010; Zhang et al. 2020). Recent publication of many genome assemblies from diverse species has resolved at least one of these problems (Genereux et al. 2020; Jebb et al. 2020; Rhie et al. 2021; Threlfall and Blaxter 2021), creating an opportunity to determine the extent of HTT.
Mammalian genomes are of considerable interest due to their propensity for relatively low TE diversity compared with most other vertebrates (Furano et al. 2004; Chalopin et al. 2015; Sotero-Caio et al. 2017), making HTT events more easily identifiable. Although typically 20–50% of mammalian genomes are TE-derived, much of this is from retrotransposons (Chalopin et al. 2015; Sotero-Caio et al. 2017); most mammals have experienced little to no DNA transposon accumulation in the last 40 million years (My) (Pace and Feschotte 2007; Sotero-Caio et al. 2017). A major exception to this observation is the order Chiroptera, especially members of the family Vespertilionidae, which are well-known for having unusually diverse TE repertoires and experiencing several recent, independent DNA transposon invasions (Pritham and Feschotte 2007; Ray et al. 2007, 2008, 2015; Thomas et al. 2011; Pagán et al. 2012; Mitra et al. 2013; Platt et al. 2016). Although the impacts of these DNA transposon invasions are not fully understood, they offer a large pool of genetic variation that may contribute to rapid genome evolution in bats. Several studies have shown that TE-driven exon shuffling and transposase co-option have impacted bat evolution (Pritham and Feschotte 2007; Thomas et al. 2014; Grabundzija et al. 2016; Cosby et al. 2021). Indeed, a fair number of DNA transposon derived genes are found in mammal and vertebrate lineages with a variety of functions including, but not limited to, transcription, chromosome structure, and immunity (reviewed in Feschotte and Pritham 2007).
Bats are the second largest order of mammals (n = 1426) (Simmons and Cirranello 2020, accessed September 4, 2021), exhibiting some of the most unique mammalian phenotypes (e.g., flight, laryngeal echolocation, extended longevity, and tolerant immunity) and inhabiting multiple ecological niches (Jebb et al. 2020). This phenotypic diversity along with their unusual diversity of younger TEs led us to investigate HT of DNA transposons involving bats. In addition to the broad array of mammalian genomes from the Zoonomia Project (Genereux et al. 2020), several bat genome assemblies have been produced by the Bat1K Project (Teeling et al. 2018; Jebb et al. 2020). Combined, this genomic data includes 37 bat species from 11 families and 28 genera spanning the 2 major chiropteran clades, Yinpterochiroptera and Yangochiroptera (Teeling et al. 2005; Amador et al. 2018). We analyzed TE accumulation patterns across Chiroptera and leveraged TE curation data from 251 mammal assemblies to perform a large-scale analysis of recent HT of DNA transposons involving bats. Our findings highlight TE-based diversity within bats and suggest that, in a radical departure from other eutherian mammals, Chiroptera is a hotspot for HTT.
Results
More Recent, Substantial DNA Transposon Accumulation in Bats
We used a curated de novo TE library to annotate TE insertions in 250 eutherian mammalian species, including 37 bat species (supplementary table S1, Supplementary Material online) (Osmanski et al. 2022; Christmas et al. forthcoming). A general comparison of TE content among mammal assemblies is available elsewhere (Osmanski et al. 2022). Rather than recapitulate that work in illustrating general distinctions between bats and nonbats, we chose eight representative eutherians as our outgroup taxa. Of the eight, four species were selected due to having the greatest accumulation of young (≤50 My) DNA transposons outside of bats: two tenrecs (Echinops telfairi and Microgale talazaci, Afrosoricida) and the Eastern mole and the Indochinese shrew (Scalopus aquaticus and Crocidura indochinensis, Eulipotyphla). The other four species along with the eulipotyphlans represent one of the five mammalian orders closely related to Chiroptera within Laurasiatheria (Foley et al. 2022): horse (Equus caballus, Perissodactyla), cow (Bos taurus, Artiodactyla), pangolin (Manis javanica, Philodota), and domestic cat (Felis catus, Carnivora).
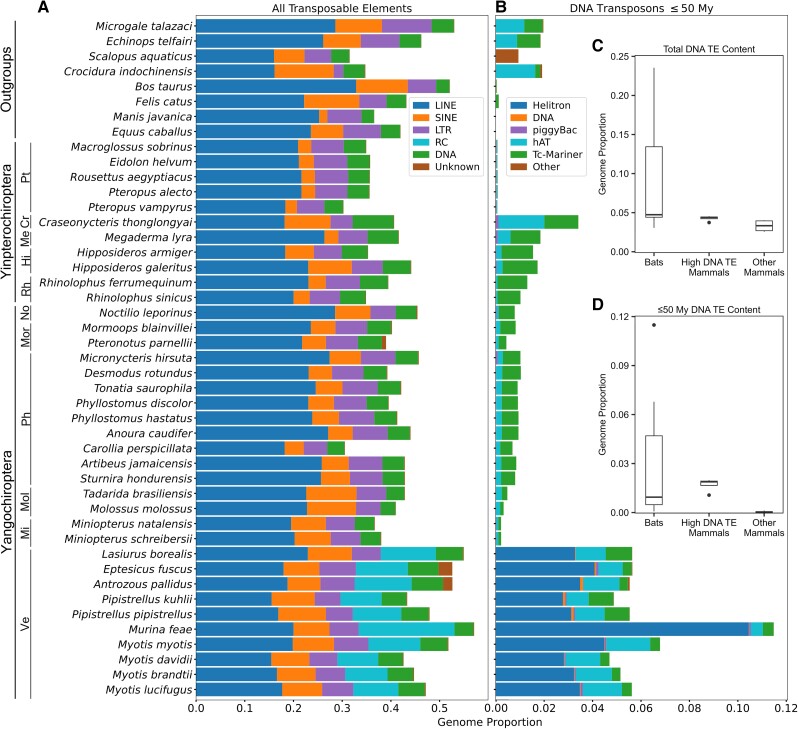
With regard to total TE content, bats generally resemble other mammals, with TEs composing 30–60% of the genome, 15–30% from LINE elements, and the rest split among SINE, LTR, and DNA elements (fig. 1A). The eight outgroup mammals are similar in proportions of different types of TEs, though the eulipotyphlans have slightly lower TE content overall, and B. taurus harbors a relatively high proportion of LINEs (Osmanski et al. 2022). The latter has been discussed previously and is due to an independent HT of RTE-like retrotransposons, Bov-B LINEs (Kordis and Gubensek 1998). Such variation in retrotransposon content is not unexpected among mammals (Sotero-Caio et al. 2017; Platt et al. 2018).
Fig. 1.
(A) Total transposable element accumulation, (B) DNA transposon accumulation within the last 50 My, and (C and D) box plots depicting ranges of total DNA transposon genome content in 37 chiropterans and 8 outgroup mammalians. High DNA TE mammals are defined as described in the main text as E. telfairi, M. talazaci, S. aquaticus, and C. indochinensis. Bat families are indicated by abbreviations left of species names and are as follows: Pt, Pteropodidae; Me, Megadermatidae; Cr, Craseonycteridae; Rh, Rhinolophidae; Hi, Hipposideridae; Ve, Vespertilionidae; Mi, Miniopteridae; Mol, Molossidae; No, Noctilionidae; Mor, Mormoopidae; and Ph, Phyllostomidae.
However, there are several major differences between bats and nonbats. Most notable is the presence of generally higher total and more recent DNA transposon accumulation (fig. 1B–D), mostly hAT and Tc-Mariner transposons, in many of the bat subclades and the obvious presence of substantial accumulation of RC elements in vespertilionid bats in the last 50 My (fig. 1B). Substantial RC accumulation is not observed in yinpterochiropteran bats or outgroup species. Within the DNA transposon categories, vespertilionid bats also have higher hAT element accumulation than yinpterochiropteran lineages, except for the bumblebee bat (Craseonycteris thonglongyai) and the lesser false vampire bat (Megaderma lyra) (fig. 1B). In comparison with nonbats, vespertilionid bats and C. thonglongyai have higher young DNA transposon accumulation than all outgroup mammals, but the four high DNA TE mammals have greater amounts of young DNA transposons than most if not all other bats. However, the other four mammals have less young DNA transposon accumulation than all bats expect pteropodids, and this low recent accumulation is more representative of eutherian mammals in general (supplementary table S2, Supplementary Material online) (Osmanski et al. 2022).
Temporal Class II Transposon Accumulation in Bats
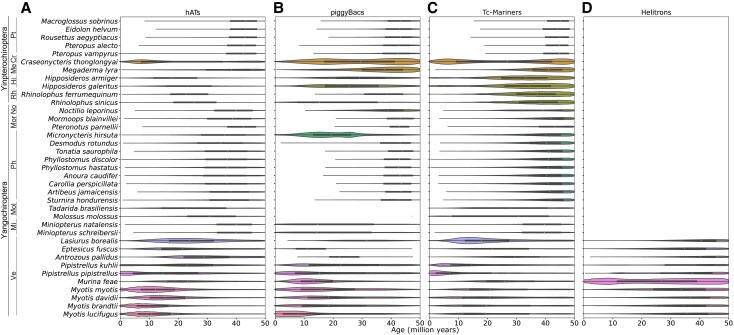
To examine the temporal context of TE accumulation, we calculated each TE copy's divergence from the TE consensus sequence and applied species-specific neutral mutation rates (supplementary table S3, Supplementary Material online) to assign insertion times to each insertion. To explore temporal variation in Class II accumulation among lineages, we visualized DNA/RC accumulation within the past ∼50 My in figure 2. This figure illustrates broad patterns of DNA transposon superfamily accumulation as it varies by bat family and patterns that are clearly lineage specific. Each superfamily comprises multiple, potentially lineage-specific subfamilies.
Fig. 2.
Violin plots of DNA transposon distributions by superfamily in bats. Distributions of (A) hAT, (B) piggyBac, (C) Tc-Mariner, and (D) Helitron elements within the last 50 My in 37 bat species. Species are arranged phylogenetically; bat families are indicated by abbreviations left of species names and are as follows: Pt, Pteropodidae; Me, Megadermatidae; Cr, Craseonycteridae; Rh, Rhinolophidae; Hi, Hipposideridae; Ve, Vespertilionidae; Mi, Miniopteridae; Mol, Molossidae; No, Noctilionidae; Mor, Mormoopidae; and Ph, Phyllostomidae.
For example, vespertilionid bats (Yangochiroptera) show substantial hAT accumulation within the last 40 My, with Myotis species showing the highest hAT accumulation between 10 and 20 Mya, coinciding with species diverging between 10.9 and 18.2 Mya (Kumar et al. 2022), whereas Lasiurus borealis appears to have experienced a slightly older peak of accumulation 20–35 Mya (fig. 2A). The two available Pipistrellus species have experienced increased hAT accumulation within the last 5 My, well after the divergence of the two species ∼9.6–17.6 Mya (Kumar et al. 2022). All vespertilionid bats show Helitron accumulation across the last 50 My, including ancestral accumulation, but Murina feae displays a surprisingly large amount, with accumulation peaks ∼10 and 40 Mya (fig. 2D). Across other yangochiropterans, Micronycteris hirsuta stands out as experiencing a burst of piggyBac accumulation not apparent in other phyllostomids (fig. 2B); otherwise, phyllostomids show consistent patterns of ancestral Tc-Mariner accumulation 40–50 Mya and little else (fig. 2). Noctilio leporinus shows high Tc-Mariner accumulation over the span of 25–50 Mya, with little accumulation more recently (fig. 2C).
Yinpterochiropterans display similarly variable Class II accumulation (fig. 2). Pteropodid bats display a uniform lack of substantial DNA transposon accumulation within the last 50 My, with little to no accumulation within the last 10 My (fig. 2). This is consistent with previous observations of no substantial retrotransposon accumulation over approximately the same period (Cantrell et al. 2008; Nikaido et al. 2020). Other yinpterochiropterans show peaks of Tc-Mariner accumulation 35–40 Mya and low-level accumulation of other DNA transposons. Craseonycteris thonglongyai and its closest relative in this study, M. lyra, both have considerably higher piggyBac accumulation and to a much lesser extent hAT accumulation than other yinpterochiropterans. However, C. thonglongyai also exhibits a striking increase of species-specific DNA transposon accumulation in the last 5–6 My, with a second peak of hAT, piggyBac, and Tc-Mariner accumulation (fig. 2A–C).
Many More HT Events in Bats Compared with Other Mammals
Lineage-specific TE subfamilies constitute much of the DNA and RC accumulation across bat lineages in the last 50 My, an observation consistent with previous studies (Pritham and Feschotte 2007; Ray et al. 2007, 2008; Thomas et al. 2011; Pagán et al. 2012; Mitra et al. 2013; Zhuo et al. 2013; Platt et al. 2016). Unlike LINE retrotransposons, which tend to accumulate over long periods and exist as multiple lineages in genomes, diversifying into sometimes numerous subfamilies (Konkel et al. 2010; Boissinot and Sookdeo 2016), DNA transposons are prone to inactivating internal deletions and tend to have shorter lifespans (Lohe et al. 1995; Smit 1996; Feschotte and Pritham 2007; Muñoz-López and García-Pérez 2010; Gilbert and Feschotte 2018). As a result, recent accumulation of a wide variety of DNA transposons is intriguing and suggests possible external origins.
Historically, the criterion used to identify a potential HTT is the presence of a unique TE in a given genome and the corresponding absence from close relatives. Although not always possible, confirming the presence of a highly similar element in the genome of a distant relative serves as strong confirmation of the HTT. An example is the presence of a piggyBac transposon, piggyBac2_ML, in the Myotis lucifugus genome, and a highly similar element, piggyBac2_Mm, in the genome of Microcebus murinus, a lemur (Pagan et al. 2010). The concurrent absence of any similar elements in the genomes of other mammals strongly suggests horizontal movement from one lineage to the other via some, usually unknown, vector, such as a virus (Gilbert et al. 2010, 2014, 2016, Gilbert and Feschotte 2018; Thomas et al. 2010).
We investigated possible HT of bat DNA transposons across mammals and other eukaryotes using a broad-scale approach (Materials and Methods). We identified 221 putative HT DNA/RC transposons representing 229 HT events involving bats (supplementary tables S4–S6, Supplementary Material online). Tc-Mariner elements are well-known as frequent participants in HT (Peccoud et al. 2017; Reiss et al. 2019; Zhang et al. 2020) and, as expected, comprise over a third of putative HT events (n = 84, 36.7%). Elements from the hAT, piggyBac, and Helitron superfamilies make up the remaining 145 HT events (n = 64, 29, 52, respectively). Blast searches indicated no copies of these putative HTTs in any available eukaryote assembly (other than the chiropteran assemblies from which it was originally detected) in all but 19 cases (see below). Previous studies (Wallau et al. 2012; Melo and Wallau 2020) have also used searches of orthologous insertion sites in addition to Blast to confirm patchy TE distributions of putative HTTs. However, the large number of mammal assemblies and putative HTTs precluded such a large number of additional searches. We therefore queried two outgroup species with high-quality genome assemblies, B. taurus and E. caballus, in detail for orthologous TE copies of the 221 putative HTTs. These searches yielded zero full-length or partial matches. These results along with the lack of Blast hits are consistent with horizontal transfer rather than prolonged vertical transmission.
Of the 19 HTTs where a nonchiropteran match was identified by Blast, 16 elements involved other eutherian clades including Lemuriformes (12 TEs), Afrosoricida (6 TEs), Scandentia (1 TE), and Eulipotyphla (1 TE) (table 1, supplementary tables S5 and S7, Supplementary Material online). These HTTs included ten hAT elements, five Tc-Mariner elements, and two piggyBac elements. Two HTTs, Mariner_Tbel and npiggy1_Mm, were previously identified as horizontal transfers involving mammals. Mariner_Tbel was previously found in the tree shrew Tupaia belangeri (Oliveira et al. 2012), consistent with our findings (supplementary tables S5 and S7, Supplementary Material online), as well as the European hedgehog Erinaceus europaeus. npiggy1_Mm, a nonautonomous piggyBac element, was previously identified as part of an HT event with its autonomous partner piggyBac1_Mm in the lemur M. murinus (Pagan et al. 2010). Zero orthologous HTT insertions were found between these mammals and bats indicating independent insertion events consistent with HT. A single autonomous hAT element, OposCharlie2, was found in a marsupial, Monodelphis domestica, consistent with previous HT studies (Gilbert et al. 2010; Novick et al. 2010). Only two elements were detected in nonmammals. An autonomous Tc-Mariner, Mariner2_pKuh, was found in an African reedfish, Erpetoichthys calabaricus, and the bat Pipistrellus kuhlii (52 and 327 copies, respectively [supplementary table S7], Supplementary material online), but not in the closely related Pipistrellus pipistrellus. This is consistent with the estimated age of the element, ∼2.2 My, which is younger than the divergence of the two pipistrelle species, ∼10–18 Mya (Kumar et al. 2022). The element has high sequence conservation as well, with 99.74% identity between the two species’ consensus sequences (supplementary fig. S1, Supplementary Material online). The second element, a nonautonomous Tc-Mariner, nMariner1_Lbo, was identified in two lizard species, Zootoca vivipara and Lacerta agilis, as well as three vespertilionid bats (supplementary table S7, Supplementary Material online), with sequence conservation of >83% among all species, > 90% excluding the single insertion in Antrozous pallidus (supplementary fig. S2, Supplementary Material online). Only 5 of the 19 putative HTTs are autonomous. Our methods assumed that many possible autonomous HTTs have <90 annotated copies in bat genomes, possibly due to loss or degradation, but that the corresponding HT events are represented by these nonautonomous counterparts.
Table 1.
Summary of Putative HT DNA Transposons Present in Multiple Eukaryote Clades.
| Number of Species Involved | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| TE Subfamily | TE Superfamily | Consensus Length (bp) | Polypteriformes | Squamata | Didelphimorphia | Afrosoricida | Scandentia | Lemuriformes | Eulipotyphla | Chiroptera |
| CraTho-1.191 | hAT | 191 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 |
| CraTho-2.327 | hAT | 213 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 3 |
| EchTel-1.100 | hAT | 334 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 5 |
| hAT-2N1_MM | hAT | 198 | 0 | 0 | 0 | 1 | 0 | 4 | 1 | 7 |
| MirCoq-4.2925 | hAT | 223 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 2 |
| MurFea-1.231 | hAT | 230 | 0 | 0 | 0 | 1 | 0 | 3 | 0 | 2 |
| MyoBra-4.2938 | hAT | 229 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 4 |
| MyoBra-5.81 | hAT | 192 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 5 |
| OposCharlie2 | hAT | 2996 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 |
| SPIN_NA_9_Ml | hAT | 311 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 5 |
| SPIN_Og | hAT | 2836 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 13 |
| npiggy1_Mm | piggyBac | 240 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| npiggBac-2_EF | piggyBac | 172 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 |
| DNA2_pKuh | Tc-Mariner | 152 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 4 |
| Mariner2_pKuh | Tc-Mariner | 2292 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| Mariner3_pKuh | Tc-Mariner | 2282 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 2 |
| Mariner_Tbel | Tc-Mariner | 1283 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 3 |
| nMar1_Rf | Tc-Mariner | 236 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 6 |
| nMariner1_Lbo | Tc-Mariner | 184 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 3 |
| Total # Unique Species | 1 | 2 | 1 | 2 | 2 | 5 | 1 | 18 | ||
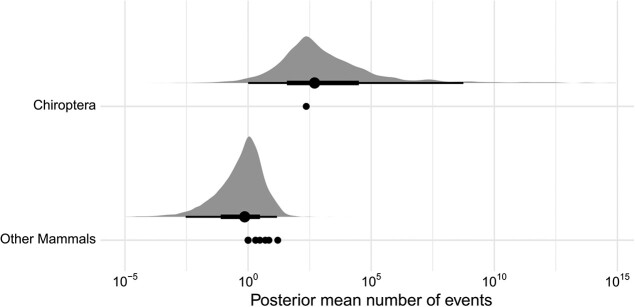
In contrast to the 229 HT events in bats, few possible HT events were identified in other mammals (detailed above and in Christmas et al. [forthcoming]). Of the 6 other orders with HT events, only Primates and Afrosoricida had more than 5 events (15 and 6, respectively). To compare HT events between the 37 bats and 213 other eutherian mammal species, we modeled the number of events by mammalian order (supplementary table S8, Supplementary Material online) using a negative binomial distribution and estimated HT means for both bats and nonbats. Although bats represent only one mammalian order, this point observation can be compared with the posterior distribution of the mean of HT events across 18 other orders (equivalent to a one sample t-test for normal data). As there is only a single order to estimate the mean for bats, posterior distribution of these estimates overlaps (fig. 3). However, considering there is only a single point estimate of HT for bats, it does not overlap with the posterior mean of HT for all other mammalian orders. This demonstrates that there were many more HT events in bats than in other mammalian orders.
Fig. 3.
Posterior distributions of group category (bats vs. nonbat eutherian mammals) on horizontal TE transfer counts. A constant of 1 was added to HTT counts for plotting to show the wide range of posterior estimates, which spans many orders of magnitude. For each coefficient: large dots show the median, thin lines show the 95% posterior probability, thick lines show the 66% posterior probability, and shaded curves show the posterior density of the estimates. Small dots show the observations on which the models were based.
Varying HT Patterns and Rates in Chiroptera
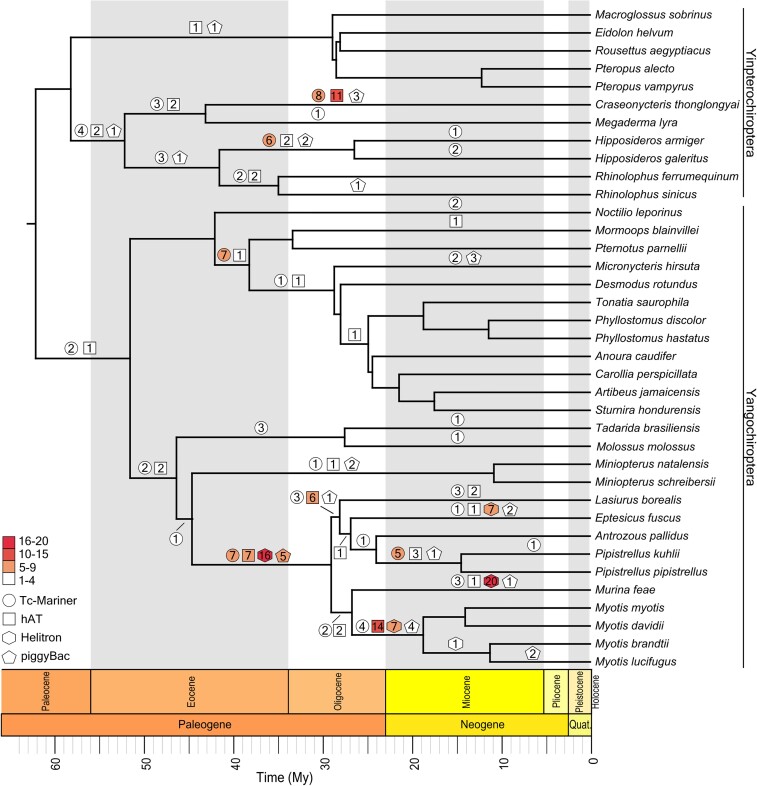
We explored large-scale patterns of HT within bats by mapping the 229 putative HT events onto a bat phylogeny based on the presence/absence patterns of each element and its estimated average age (fig. 4, supplementary table S6, Supplementary Material online). As expected, there were far more putative HT events in yangochiropterans than in yinpterochiropteran lineages (170 and 59, respectively), but the distribution is exceptionally uneven within each clade. More than a third of all HT in Yinpterochiroptera are unique to C. thonglongyai, with only two relatively ancient examples occurring in Pteropodidae. Similarly, within Yangochiroptera, a large majority (n = 134, 78.8%) of HT events involve only vespertilionid bats. Interestingly, eight different elements appear to have independently invaded both Vespertilionidae and either C. thonglongyai or the Rhinolophoidea ancestral branch (supplementary tables S5 and S6, Supplementary Material online), though it is unclear if these represent initial HT into one bat clade followed by HT between bats, a pair of independent HT from outside Chiroptera into different bat clades, or some combination thereof. Searches for orthologous insertions of the eight HTTs among representative species (Hipposideros galeritus, C. thonglongyai, Myotis myotis, and P. pipistrellus) yielded zero matching orthologous insertions.
Fig. 4.
Horizontal transfer of DNA transposons within Chiroptera. Inferred horizontal transfer (HT) events of 221 unique TEs from Tc-Mariner (circle), hAT (square), Helitron (hexagon), or piggyBac (pentagon) superfamilies are labeled on corresponding branches. Shape color indicates numerical range of putative HT events of a given branch: white, 1–4 elements; pink, 5–9; red, 10–15; and dark red, 16–20; the number of events was included within each marker. Phylogeny is scaled by estimated divergence times in millions of years (My). HT event branch assignment was inferred from presence/absence patterns and the element's average age. Phylogenetic relationships are based on Foley et al. (2022) and Amador et al. (2018); estimated divergence times were (supplementary table S10, Supplementary Material online) taken from TimeTree (Kumar et al. 2022).
We then calculated HT event rates for bat lineages. Yangochiropterans had almost double the average HT rate of yinpterochiropterans, with a rate of 0.277 versus 0.146 putative HT/My, respectively (supplementary table S9, Supplementary Material online). However, we found a broad range of HT rates within both groups. Within Yinpterochiroptera, rates ranged from 0.023 for M. lyra to 0.512 for C. thonglongyai. The ancestral branch for Hipposideridae and Rhinolophidae had the second-highest rate at 0.244. Within Yangochiroptera, rates varied between 0.022 at the ancestral branch for Miniopteridae and Vespertilionidae and 1.593 at the ancestral branch for the four Myotis species, which was also the highest HT rate within examined lineages. The second-highest rate within Yangochiroptera was in the ancestral Vespertilionidae branch, at 1.215 (supplementary table S9, Supplementary Material online).
Within bats, Hipposideridae, Rhinolophidae, and Vespertilionidae are among the most species-rich clades while also exhibiting some of the highest TE diversity. This raises the question of a relationship between species richness and HT events. The relationship between species diversity and HT events was indeed stronger than for mammals more generally (fig. 1). However, the relationship between species richness and HT events, despite considerable variation across TE types, proved to be statistically unsupported (supplementary fig. S4 and table S11, Supplementary Material online) but intriguing. This was also the case for only young TE counts (supplementary fig. S5 and table S12, Supplementary Material online). By increasing statistical power, additional data have the potential to influence future understanding of this relationship.
Discussion
Our results, in combination with those of Christmas et al. (forthcoming), indicate that bats are a hotspot for horizontal transfer of DNA transposons within mammals. This was a broad-scale, computational approach to identify HTT, and we used several conservative search thresholds that excluded candidate HT DNA transposons with low copy number (<90 annotated insertions) in bats, such as Helibat1 and SPIN_Ml, both previously identified as HTT with limited distributions (Pace et al. 2008; Thomas et al. 2011). We also excluded many highly similar elements to avoid inflation from vertically diversifying elements, including highly similar deletion products. This could have yielded false negatives in both our mammalian targets and other eukaryotes. Further research into potential vectors such as eukaryotic parasites and viruses will require less conservative methods to detect low copy or fragmented elements. Despite these limitations, we found several hundred HT events, which likely are an underrepresentation of the number of HT events that have occurred within Chiroptera, particularly as HT is more likely than vertical persistence of DNA transposons (reviewed in Feschotte and Pritham 2007; Wells and Feschotte 2020). In comparison with other mammals, bats have far more HT events and substantially higher recent DNA transposon accumulation, even when compared with mammals known to have experienced HTT, such as Otolemur garnettii, M. murinus, or E. telfairi (figs. 1 and 3, supplementary tables S2 and S8, Supplementary Material online). Although our searches did identify four species with higher than the mammalian average recent DNA transposon accumulation, these instances are clearly exceptions among nonbat eutherians and not the rule.
To better clarify the distributions and impacts of these HT events, more even sampling across bat lineages is required, particularly within large species complexes. For example, the genus Rhinolophus consists of ∼100 species divided among 15 species groups (Csorba et al. 2003; Stoffberg et al. 2010; Demos et al. 2019), but was represented by only two genome assemblies. Since most genera are only represented by a single species, it should be noted that HT events mapped to terminal branches may represent HTs into a common ancestor of multiple species rather than our representative terminal species. That said, underrepresentation within genera would not explain the numerous lineage-specific HTs of C. thonglongyai (26), which is a monotypic genus.
Consistent with the TE-Thrust hypothesis, most inferred HT events in figure 4 map to families or genera that have undergone rapid diversification. Owing to their potential for genomic innovation, TE expansions in a genome represent an opportunity for those genomes to gain variation that could lead to adaptive opportunities (Oliver and Greene 2011, 2012), giving rise to the TE-Thrust hypothesis. HT events are concentrated at the base of Hipposideros and Rhinolophus (Foley et al. 2015), which have 90 and 106 recognized species, respectively, and Vespertilionidae (Lack and Van Den Bussche 2010), which currently consists of 512 species, and basal lineages within it, such as genus Myotis, which comprises 131 species (Simmons and Cirranello 2020, accessed September 4, 2021). Thus, intermittent HT and subsequent bursts of TE amplification correspond to diversification of several large clades across Chiroptera. The TE-thrust hypothesis also proposes a “Goldilocks Zone” of TEs and evolutionary potential: Too little TE activity results in evolutionary stasis, and too much would cause detrimental genomic instability, but moderate amounts of TE activity and accumulation can allow genomic dynamism and potentially rapid lineage evolution and diversification (Oliver and Greene 2011, 2012). The data we present are consistent with these predictions. Some bat lineages, having experienced an influx of highly successful DNA transposons, may have exploited the increased genomic diversity to aid their expansion into multiple niches. Alternatively, higher species richness could lead to more HT events due to increased ecological interactions with potential HT sources and/or vectors, which could synergize with initial HT-driven diversification. Or environmental heterogeneity may promote speciation and HT, without HT directly impacting species diversification. This seems less likely given documented Helitron capture of host promoters and exons in Myotis (Thomas et al. 2014). Helitron-driven tissue-specific nuclear gene transcription was shown in Myotis brandtii (Grabundzija et al. 2016), and Cosby et al. (2021) identified numerous DNA transposase–gene fusions with broad gene regulatory functions that vary across bat clades, including two fusion genes specific to vespertilionids. However, we did not find statistical support for associations between HT elements and descendent species richness or young (≤50 My) TE accumulation and species richness, likely from underestimating diversity within DNA transposon superfamilies and due to the few bat species sampled and the high variance of species richness represented by each of our focal taxa. We plan to address this in the future as additional high-quality genome assemblies are released and statistical power is increased.
Although we do not know why bats are hotspots for HT and HT-associated TE diversity and accumulation, our results may indicate a higher tolerance for TE activity in bats. Possible factors influencing this presumed tolerance could include adaptations in DNA repair pathways and expression (Seim et al. 2013; Zhang et al. 2013; Foley et al. 2018; Huang et al. 2019) allowing higher TE loads. Tolerance may also have been influenced by the potential adaptations in bat immune responses that allow them to experience low viral loads but many circulating viruses with little apparent negative effects and rapid viral spreading in hosts (Subudhi et al. 2019; Brook et al. 2020; Jebb et al. 2020; Irving et al. 2021; Moreno Santillán et al. 2021). As viruses are likely candidates for transferring TEs (Gilbert et al. 2010, 2014, 2016; Thomas et al. 2010; Gilbert and Feschotte 2018), variability within and across bat lineages in these immune-related gene expansions and losses (Moreno Santillán et al. 2021), diversity of viruses present (Jebb et al. 2020), as well as impacts of variable geographic proximity (Peccoud et al. 2017) may help explain the higher frequency of HTT in chiropterans and variability of HT success across bat lineages.
Differential bat ecology may also represent part of the answer. Previous studies have implicated blood feeding arthropods such as Rhodnius prolixus, an insect vector of Chagas disease, as a vector for HT (Gilbert et al. 2010; Matthews et al. 2011). Herbivorous bats have significantly less recent DNA transposon accumulation than carnivorous species (Osmanski et al. 2022). These observations suggest insectivorous species may be more susceptible to HT than species with other dietary habits. And indeed, the clade of bats exhibiting the highest rate of putative HT in our study is the family Vespertilionidae, which is almost exclusively insectivorous (Nowak 1999; Fenton and Bogdanowicz 2002; Morales et al. 2019). Craseonycteris thonglongyai, rhinolophids, and hipposiderids are also insectivorous (Arbour et al. 2019; Pavey 2021) and stand out as exceptional genomic habitats for HT of DNA transposons. Yet despite their openness to HT, only a handful of types have been successful, and with the emphatic exception of Helitrons in vespertilionids, bats do not seem to have much more diversity in DNA transposons compared with other eutherians. Why this is the case is still unclear.
The potential impacts of these HTT on bat genome evolution cannot be understated. TEs generally are a potent source of genomic variation that can impact genes and genome structure in numerous ways (Schaack et al. 2010; Oliver and Greene 2012; Casacuberta and González 2013; Gilbert and Feschotte 2018). Studies in other mammals have shown low conservation of regulatory sites, and TEs play critical roles in restructuring regulatory networks by contributing lineage-specific transcription factor binding sites and regulatory elements (Wang et al. 2007; Kunarso et al. 2010; Schmidt et al. 2012; Chuong et al. 2013; Jacques et al. 2013; Sundaram et al. 2014; Notwell et al. 2015; Trizzino et al. 2017; Judd et al. 2021). DNA transposons are no exception. Previous work has shown Helitron-mediated exon and promoter shuffling and substantial genome inflation within bats (Thomas et al. 2014), as well as transposon co-option events resulting in gene fusion and changes in gene network regulation (Cosby et al. 2021). DNA transposons are well suited to exaptation into transcription factors, as their encoded transposase proteins, a DNA binding domain and a catalytic nuclease domain, can be domesticated or repurposed for host cellular functions (Feschotte and Pritham 2007). Known host–transposase fusion genes include GTF2IRD2 in placental mammals (Tipney et al. 2004), SETMAR and CSB-PGBD3 in primates (Cordaux et al. 2006; Newman et al. 2008), and KRABINER in Vespertilionid bats (Cosby et al. 2021).
We note that a weakness of our study is the identification of only a few potential donor/recipient relationships to the species level. This, however, is to be expected given the paucity of animal genome assemblies available to search. Only several thousand animal genomes are available of the ∼7.8 million animal species currently estimated to exist (Mora et al. 2011). Thus, although determining the likely HT partner in any given HT event would be ideal, doing so in all cases is difficult. We point out that, given our current understanding of evolutionary processes, the sudden appearance of multiple intact sequences with the hallmarks of DNA transposons in a lineage is likely the result of HT.
The observations presented here suggest that HTT events involving Class II TEs contribute to bat genomic diversity to a degree not found in other mammals. The cause of this propensity toward DNA transposon invasion is currently a mystery, but future investigations may reveal the genomic characteristics that make one species more or less likely to be a safe harbor for HT TEs. Regardless of the reasons and mechanisms behind the multiple invasions, the correspondence between high rates of HTT events and species radiations in several large bat clades suggests that HTT activity facilitates genomic innovation and taxonomic diversity. Our results shed new light on the extent of HTT in bats, but not the impacts of each example or lineage. More research is needed to clarify the specific roles that these TE expansions have played in bat diversification and genome evolution.
Materials and Methods
Taxon Selection
We examined 37 bat genome assemblies and 214 other eutherian mammal assemblies for this work (supplementary table S1, Supplementary Material online). These included assemblies from the Zoonomia sequencing effort (Genereux et al. 2020), publicly available assemblies, and from other sources such as the Bat1k consortium (Jebb et al. 2020; Wang et al. 2020; Moreno Santillán et al. 2021). In cases where species were represented by individuals in the Zoonomia project, but the assemblies generated by other efforts were of higher quality, we replaced the Zoonomia assemblies with the alternates (supplementary table S13, Supplementary Material online). We used a combination of PacBio, Bionano, HiC, and Illumina sequencing to generate high-quality assemblies for Eptesicus fuscus and A. pallidus (see supplementary Methods, Supplementary Material online).
Annotation of Mammalian Transposable Element Insertions
We used the curated de novo transposable element (TE) consensus sequence library described in Osmanski et al. (2022) to annotate TE insertions in all selected species using RepeatMasker v4.1.2-p1 (Smit et al. 2013-2015) with the RMBlast search engine. Output was processed using RM2Bed.py, a utility in the RepeatMasker package, with TE insertion overlap resolution by lower divergence values (-o lower_div). TE insertion accumulation and temporal distributions were visualized using matplotlib (Hunter 2007) in Python v3.7.6. We estimated individual TE insertion ages by calculating species-specific neutral mutation rates for all lineages within the last ∼50 My using pairwise branch lengths from Foley et al. (2022) and median divergence times for each species versus an outgroup mammal taxon from TimeTree (Kumar et al. 2022). We then evaluated the TE content of the 213 nonbat eutherian mammals and selected the 4 species with the highest recent DNA transposon accumulation to compare to bats, as well as 4 other species representing eutherian orders closely related to Chiroptera. Annotations for rolling-circle elements (Helitrons) in bat species outside of Vespertilionidae were excluded from these visualizations, as these are known to be false positives, as discussed in Osmanski et al. (2022).
Identification of Putative HT Class II TEs Involving Chiroptera and Other Mammals
We selected DNA/RC elements with ≥90 annotated copies in at least 1 bat species as our initial set of HT candidates. We then used the library consensus sequences (107) of this initial TE set as queries in Blast searches utilizing what we refer to as the 90-90-90 rule (described below), a more conservative version of the 80-80-80 rule developed by Wicker et al. (Wicker et al. 2007). We searched for TE copies meeting our conservative criteria of present in the genome assemblies of one or more bat species. To identify any additional eukaryote involvement, we performed Blast searches of these elements across all available eukaryote genome assemblies in the NCBI databases.
Putative HTT were defined as TE insertions annotated in an assembly with <90 insertions called in closely related species. We narrowed our search for HTT to DNA transposon and rolling-circle transposons with ≥90 copies annotated by RepeatMasker in one or more bat species. We then used the same TE consensus sequences as queries for BlastN searches (Blast + v2.11.0 [Camacho et al. 2009]) in the said bat genomes and implemented the 90-90-90 rule to identify potential HTTs. The criteria of the 90-90-90 rule are 1) the element must be ≥90 bp in length, 2) share ≥90% sequence identity with one another, and 3) have a total ungapped length matching ≥90% of the consensus sequence. To further exclude potentially erroneous hits from similar elements harboring short insertions, the element copies must have been ≤10% longer than the query consensus sequence length. We also excluded potential duplicate elements or vertically diversifying elements with ≤5% sequence divergence using the cross_match utility of Phrap v0.990319 (Gordon 2003). Similarly, to account for and exclude DNA transposon deletion products, we used the same query consensus sequences as before to perform a modified CD-HIT (Storer, Hubley, Rosen and Smit 2021) search for candidate HTT sequences that cluster together. This search performs two successive cd-hit searches. The first clusters elements ≥ 90% identical, and the second search adds elements >80% similar to existing clusters or generates new ones. Elements that clustered together and had overlapping presence/absence patterns across bat species were collapsed into a single presumed HT event.
We then performed a final manual curation by comparing alignments of candidate HTT consensus sequences with all other elements in the TE consensus library from Annotation of Mammalian Transposable Element Insertions to identify any deletion products that were not identified in the previous clustering step. To estimate the age of each TE insertion within a species, we calculated modified Kimura two-parameter (K2P) distances for each TE copy compared with the library consensus sequence using RepeatMasker's alignAndCallConsensus and Linup utilities (Smit et al. 2013-2015). We then mapped the HT events onto a phylogenetic tree of our 37 bat species based on the presence/absence pattern of the putative HT elements from our filtered BlastN results and their average ages. TE ages were calculated per species using the average K2P distance and the species-specific neutral mutation rates. The phylogenetic tree was built based on Foley et al. (2022) and Amador et al. (2018) and used a combination of nonconflicting average or median divergence estimates from TimeTree (supplementary table S10, Supplementary Material online) (Kumar et al. 2022), accessed September 3, 2021.
Orthologous TE Insertion Searches within Mammalia
To identify possible orthologous copies of putative HTTs, we performed pairwise orthologous site searches between 28 bats species and 2 mammal outgroups, B. taurus and E. caballus, using Zoonomia's 241 mammal genome alignment (Genereux et al. 2020). With the exception of N. leporinus, the other eight bat species not present in the genome alignment were represented by other members in the same family, if not the same genus. For each of the 28 bat species, we generated a BED file of the coordinates of each copy of a putative HTT in the final data set from Identification of Putative HT Class II TEs Involving Chiroptera and Other Mammals with 50 bp flanking sequence on either end. We then identified the orthologous sections of the outgroup genomes with the utility halLiftover and merged all close (≤2 bp) coordinate hits for the same TE copy into a single hit using BEDTools sort and mergeBed (Quinlan and Hall 2010). We then performed a series of TE annotations for all orthologous sites in the target outgroup species, first using RepeatMasker (Smit et al. 2013-2015) with a combined mammalian TE consensus library of ancestral mammal repeats from the Dfam database v.3.6 (Storer, Hubley, Rosen, Wheeler et al. 2021) and our original library from Annotation of Mammalian Transposable Element Insertions. Any annotations matching 1 of the 221 putative HTTs were then subjected to an additional annotation and alignment with the cross_match utility (Gordon 2003). Any cross_match annotations matching 1 of the 221 putative HTTs were then manually checked for 1) TE identity match to the copy at the bat site, 2) alignment size and score, and 3) site alignment to bat species (e.g., where there were large [>1,000 bp] gaps). The same process of pairwise orthologous site searches was performed with representative species for mammal groups harboring any of putative HTTs, which included M. talazaci (Afrosoricida), Tupaia chinensis (Scandentia), Nycticebus coucang (Lemuriformes), and C. indochinensis (Eulipotyphla). These mammals were paired with representative bat species: H. galeritus, M. myotis, M. feae, and/or P. pipistrellus. We also performed orthologous site searches between representatives of the two bat suborders: Yinpterochiroptera (C. thonglongyai, H. galeritus) and Yangochiroptera (M. myotis, P. pipistrellus).
Identification of Putative HT TEs outside of Mammalia
After identifying HT events, we applied the above methodology to identify possible HT events between Chiroptera and nonmammal eukaryotes. We performed BlastN searches of the eukaryotic reference genome database (accessed April 6, 2021 [Camacho et al. 2009]), excluding mammals, using the consensus sequences from the putative chiropteran HTTs as our query input. To reduce false negatives in distantly related taxa, we used the criterion of ≥90 full-length or near full-length copies for nonautonomous elements and a lower threshold of ≥50 copies for autonomous elements. As nonautonomous copies tend to make up the majority of DNA transposon insertions (Lohe et al. 1995; Feschotte and Pritham 2007; Muñoz-López and García-Pérez 2010), this threshold is more likely to detect true evolutionarily recent HTT in more distantly related organisms. To identify autonomous elements, we searched for open reading frames (ORFs) via the getorf utility of EMBOSS v6.6.0 (Rice et al. 2000) in species-specific consensus sequences of the putative HTT generated from a custom script, extend_align.sh, which is available on GitHub (https://github.com/davidaray/bioinfo_tools). We identified transposase ORFs by performing BlastX searches.
Testing for Associations with Species Richness
Two sets of analyses were conducted. First, we tested the association between HT TEs and fraction species richness modeling both these variables with errors. Then, we modeled fraction species richness as a function of cumulative young (≤50 My) TEs (see supplementary Methods, Supplementary Material online for details).
Supplementary Material
Acknowledgments
This project was supported by the National Science Foundation (grant numbers DEB 1838283 and IOS 2032006 to D.M.-S. and Dav.R. and DEB 1838273 and DGE 1633299 to L.D.), National Institutes of Health (grant numbers R01HG002939 and U24HG010136 to J.S., R.H., A.F.A.S., and Je.R.), NHGRI (grant number R01HG008742 to Z.C.), Irish Research Council (grant number IRCLA/2017/58 to E.T.), Science Foundation Ireland (grant number 19/FFP/6790 to E.T.), Max Planck Research Group awarded by the Max Planck Gesellschaft to S.V., Human Frontiers Science Program (grant number RGP0058/2016 to S.V.), UK Research and Innovation (grant number MR/T021985/1 to S.V.), and the Swedish Research Council Distinguished Professor Award to K.L.-T. The High-Performance Computing Center at Texas Tech University and the SeaWulf computing system at Stony Brook University provided computational infrastructure throughout the work.
Contributor Information
Nicole S Paulat, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Jessica M Storer, Institute for Systems Biology, Seattle, WA.
Diana D Moreno-Santillán, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Austin B Osmanski, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Kevin A M Sullivan, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Jenna R Grimshaw, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Jennifer Korstian, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Michaela Halsey, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Carlos J Garcia, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Claudia Crookshanks, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Jaquelyn Roberts, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Arian F A Smit, Institute for Systems Biology, Seattle, WA.
Robert Hubley, Institute for Systems Biology, Seattle, WA.
Jeb Rosen, Institute for Systems Biology, Seattle, WA.
Emma C Teeling, School of Biology and Environmental Science, University College Dublin, Belfield, Dublin 4, Ireland.
Sonja C Vernes, Neurogenetics of Vocal Communication Group, Max Planck Institute for Psycholinguistics, Nijmegen, The Netherlands; Donders Institute for Brain, Cognition and Behaviour, Nijmegen, The Netherlands; School of Biology, The University of St Andrews, Fife, United Kingdom.
Eugene Myers, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany.
Martin Pippel, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany.
Thomas Brown, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany.
Michael Hiller, LOEWE Centre for Translational Biodiversity Genomics, Frankfurt, Germany.
Danny Rojas, Department of Natural Sciences and Mathematics, Pontificia Universidad Javeriana Cali, Valle del Cauca, Colombia.
Liliana M Dávalos, Department of Ecology and Evolution, Stony Brook University, Stony Brook, NY; Consortium for Inter-Disciplinary Environmental Research, Stony Brook University, Stony Brook, NY.
Kerstin Lindblad-Toh, Science for Life Laboratory, Department of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden; Broad Institute of MIT and Harvard, Cambridge, MA.
Elinor K Karlsson, Broad Institute of MIT and Harvard, Cambridge, MA; Program in Bioinformatics and Integrative Biology, UMass Chan Medical School, Worcester, MA; Program in Molecular Medicine, UMass Chan Medical School, Worcester, MA.
David A Ray, Department of Biological Sciences, Texas Tech University, Lubbock, TX.
Zoonomia Consortium:
Gregory Andrews, Joel C Armstrong, Matteo Bianchi, Bruce W Birren, Kevin R Bredemeyer, Ana M Breit, Matthew J Christmas, Hiram Clawson, Joana Damas, Federica Di Palma, Mark Diekhans, Michael X Dong, Eduardo Eizirik, Kaili Fan, Cornelia Fanter, Nicole M Foley, Karin Forsberg-Nilsson, Carlos J Garcia, John Gatesy, Steven Gazal, Diane P Genereux, Linda Goodman, Jenna Grimshaw, Michaela K Halsey, Andrew J Harris, Glenn Hickey, Michael Hiller, Allyson G Hindle, Robert M Hubley, Graham M Hughes, Jeremy Johnson, David Juan, Irene M Kaplow, Elinor K Karlsson, Kathleen C Keough, Bogdan Kirilenko, Klaus-Peter Koepfli, Jennifer M Korstian, Amanda Kowalczyk, Sergey V Kozyrev, Alyssa J Lawler, Colleen Lawless, Thomas Lehmann, Danielle L Levesque, Harris A Lewin, Xue Li, Abigail Lind, Kerstin Lindblad-Toh, Ava Mackay-Smith, Voichita D Marinescu, Tomas Marques-Bonet, Victor C Mason, Jennifer R S Meadows, Wynn K Meyer, Jill E Moore, Lucas R Moreira, Diana D Moreno-Santillan, Kathleen M Morrill, Gerard Muntané, William J Murphy, Arcadi Navarro, Martin Nweeia, Sylvia Ortmann, Austin Osmanski, Benedict Paten, Nicole S Paulat, Andreas R Pfenning, BaDoi N Phan, Katherine S Pollard, Henry E Pratt, David A Ray, Steven K Reilly, Jeb R Rosen, Irina Ruf, Louise Ryan, Oliver A Ryder, Pardis C Sabeti, Daniel E Schäffer, Aitor Serres, Beth Shapiro, Arian F A Smit, Mark Springer, Chaitanya Srinivasan, Cynthia Steiner, Jessica M Storer, Kevin A M Sullivan, Patrick F Sullivan, Elisabeth Sundström, Megan A Supple, Ross Swofford, Joy-El Talbot, Emma Teeling, Jason Turner-Maier, Alejandro Valenzuela, Franziska Wagner, Ola Wallerman, Chao Wang, Juehan Wang, Zhiping Weng, Aryn P Wilder, Morgan E Wirthlin, James R Xue, and Xiaomeng Zhang
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Author Contributions
N.P. and Dav.R. contributed to conceptualization, design, and data analysis and interpretation and drafted the manuscript. N.P., J.S., A.O., K.A.M.S., J.K., J.G., Michaela H., C.G., C.C., Ja.R., Je.R., R.H., A.F.A.S., and Dav.R. participated in library validation and curation. Genome assembly was accomplished by M.P., T.B., and Michael H. Methods and interpretation were contributed by N.P., J.S., A.O., Dav.R., L.D., Dan.R., K.L.-T., E.K., and D.M.-S. All authors contributed to review and editing of the final manuscript. All authors gave final approval and agreed to be accountable for all aspects of the work.
Data Availability
All assemblies are available on GenBank (see supplementary table S1 , Supplementary material online for accession numbers). TE consensus sequences, and their seed alignments, are available via the Dfam database. All other data is available in the supplementary materials, Supplementary Material online; code used in the analysis is available at github.com/daray/bat_ht.
Conflict of interest statement. The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.
References
- Amador LI, Moyers Arévalo RL, Almeida FC, Catalano SA, Giannini NP. 2018. Bat systematics in the light of unconstrained analyses of a comprehensive molecular supermatrix. J Mammal Evol. 25:37–70. [Google Scholar]
- Arbour JH, Curtis AA, Santana SE. 2019. Signatures of echolocation and dietary ecology in the adaptive evolution of skull shape in bats. Nat Commun. 10:2036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boissinot S, Sookdeo A. 2016. The evolution of LINE-1 in vertebrates. Genome Biol Evol. 8:3485–3507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook CE, Boots M, Chandran K, Dobson AP, Drosten C, Graham AL, Grenfell BT, Müller MA, Ng M, Wang LF, et al. 2020. Accelerated viral dynamics in bat cell lines, with implications for zoonotic emergence. Elife 9:e48401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantrell MA, Scott L, Brown CJ, Martinez AR, Wichman HA. 2008. Loss of LINE-1 activity in the megabats. Genetics 178:393–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casacuberta E, González J. 2013. The impact of transposable elements in environmental adaptation. Mol Ecol. 22:1503–1517. [DOI] [PubMed] [Google Scholar]
- Chalopin D, Naville M, Plard F, Galiana D, Volff J-N. 2015. Comparative analysis of transposable elements highlights mobilome diversity and evolution in vertebrates. Genome Biol Evol. 7:567–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christmas MJ, Kaplow IM, Genereux DP, Dong MX, Hughes GM, Li X, Sullivan PF, Hindle AG, Andrews G, Armstrong JC, et al. forthcoming. Evolutionary constraint and innovation across hundreds of placental mammals. Science [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chuong EB, Rumi MA, Soares MJ, Baker JC. 2013. Endogenous retroviruses function as species-specific enhancer elements in the placenta. Nat Genet. 45:325–329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA. 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet. 10:691–703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Udit S, Batzer MA, Feschotte C. 2006. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc Natl Acad Sci USA 103:8101–8106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cosby RL, Judd J, Zhang R, Zhong A, Garry N, Pritham EJ, Feschotte C. 2021. Recurrent evolution of vertebrate transcription factors by transposase capture. Science 371:eabc6405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csorba G, Ujhelyi P, Thomas N. 2003. Horseshoe bats of the world (Chiroptera: Rhinolophidae). Shropshire: Alana Books. [Google Scholar]
- Demos TC, Webala PW, Goodman SM, Kerbis Peterhans JC, Bartonjo M, Patterson BD. 2019. Molecular phylogenetics of the African horseshoe bats (Chiroptera: Rhinolophidae): expanded geographic and taxonomic sampling of the Afrotropics. BMC Evol Biol. 19:166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Baidouri M, Carpentier M-C, Cooke R, Gao D, Lasserre E, Llauro C, Mirouze M, Picault N, Jackson SA, Panaud O. 2014. Widespread and frequent horizontal transfers of transposable elements in plants. Genome Res. 24:831–838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton MB, Bogdanowicz W. 2002. Relationships between external morphology and foraging behaviour: bats in the genus Myotis. Can J Zool. 80:1004–1013. [Google Scholar]
- Feschotte C. 2008. Transposable elements and the evolution of regulatory networks. Nat Rev Genet. 9:397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. 2007. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 41:331–368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, Hughes GM, Huang Z, Clarke M, Jebb D, Whelan CV, Petit EJ, Touzalin F, Farcy O, Jones G, et al. 2018. Growing old, yet staying young: the role of telomeres in bats’ exceptional longevity. Sci Adv. 4:eaao0926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, Mason VC, Harris AJ, Bredemeyer KR, Damas J, Lewin HA, Eizirik E, Gatesy J, Springer MS, Murphy WJ. 2022. A genomic timescale for placental mammal evolution. bioRxiv. 2022.2008.2010.503388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foley NM, Thong VD, Soisook P, Goodman SM, Armstrong KN, Jacobs DS, Puechmaille SJ, Teeling EC. 2015. How and why overcome the impediments to resolution: lessons from rhinolophid and hipposiderid bats. Mol Biol Evol. 32:313–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furano AV, Duvernell DD, Boissinot S. 2004. L1 (LINE-1) retrotransposon diversity differs dramatically between mammals and fish. Trends Genet. 20:9–14. [DOI] [PubMed] [Google Scholar]
- Genereux DP, Serres A, Armstrong J, Johnson J, Marinescu VD, Murén E, Juan D, Bejerano G, Casewell NR, Chemnick LG, et al. 2020. A comparative genomics multitool for scientific discovery and conservation. Nature 587:240–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Chateigner A, Ernenwein L, Barbe V, Bézier A, Herniou EA, Cordaux R. 2014. Population genomics supports baculoviruses as vectors of horizontal transfer of insect transposons. Nat Commun. 5:3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Feschotte C. 2018. Horizontal acquisition of transposable elements and viral sequences: patterns and consequences. Curr Opin Genet Dev. 49:15–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Peccoud J, Chateigner A, Moumen B, Cordaux R, Herniou EA. 2016. Continuous influx of genetic material from host to virus populations. PLoS Genet. 12:e1005838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert C, Schaack S, Pace Ii JK, Brindley PJ, Feschotte C. 2010. A role for host–parasite interactions in the horizontal transfer of transposons across phyla. Nature 464:1347–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon D. 2003. Viewing and editing assembled sequences using Consed. Curr Protoc Bioinformatics Chapter. 11:Unit11.12. [DOI] [PubMed] [Google Scholar]
- Grabundzija I, Messing SA, Thomas J, Cosby RL, Bilic I, Miskey C, Gogol-Döring A, Kapitonov V, Diem T, Dalda A, et al. 2016. A Helitron transposon reconstructed from bats reveals a novel mechanism of genome shuffling in eukaryotes. Nat Commun. 7:10716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Z, Whelan CV, Foley NM, Jebb D, Touzalin F, Petit EJ, Puechmaille SJ, Teeling EC. 2019. Longitudinal comparative transcriptomics reveals unique mechanisms underlying extended healthspan in bats. Nat Ecol Evol. 3:1110–1120. [DOI] [PubMed] [Google Scholar]
- Hunter JD. 2007. Matplotlib: a 2D graphics environment. Comput Sci Eng. 9:90–95. [Google Scholar]
- Irving AT, Ahn M, Goh G, Anderson DE, Wang LF. 2021. Lessons from the host defences of bats, a unique viral reservoir. Nature 589:363–370. [DOI] [PubMed] [Google Scholar]
- Jacques P, Jeyakani J, Bourque G. 2013. The majority of primate-specific regulatory sequences are derived from transposable elements. PLoS Genet. 9:e1003504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jebb D, Huang Z, Pippel M, Hughes GM, Lavrichenko K, Devanna P, Winkler S, Jermiin LS, Skirmuntt EC, Katzourakis A, et al. 2020. Six reference-quality genomes reveal evolution of bat adaptations. Nature 583:578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Judd J, Sanderson H, Feschotte C. 2021. Evolution of mouse circadian enhancers from transposable elements. Genome Biol. 22:193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kofler R, Senti K-A, Nolte V, Tobler R, Schlötterer C. 2018. Molecular dissection of a natural transposable element invasion. Genome Res. 28:824–835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konkel MK, Walker JA, Batzer MA. 2010. LINEs and SINEs of primate evolution. Evol Anthropol. 19:236–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordis D, Gubensek F. 1998. Unusual horizontal transfer of a long interspersed nuclear element between distant vertebrate classes. Proc Natl Acad Sci USA. 95:10704–10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Suleski M, Craig JM, Kasprowicz AE, Sanderford M, Li M, Stecher G, Hedges SB. 2022. TimeTree 5: an expanded resource for species divergence times. Mol Biol Evol. 39:msac174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunarso G, Chia NY, Jeyakani J, Hwang C, Lu X, Chan YS, Ng HH, Bourque G. 2010. Transposable elements have rewired the core regulatory network of human embryonic stem cells. Nat Genet. 42:631–634. [DOI] [PubMed] [Google Scholar]
- Lack JB, Van Den Bussche RA. 2010. Identifying the confounding factors in resolving phylogenetic relationships in Vespertilionidae. J Mammal. 91:1435–1448. [Google Scholar]
- Lampe DJ, Churchill ME, Robertson HM. 1996. A purified mariner transposase is sufficient to mediate transposition in vitro. EMBO J. 15:5470–5479. [PMC free article] [PubMed] [Google Scholar]
- Lohe AR, Moriyama EN, Lidholm DA, Hartl DL. 1995. Horizontal transmission, vertical inactivation, and stochastic loss of mariner-like transposable elements. Mol Biol Evol. 12:62–72. [DOI] [PubMed] [Google Scholar]
- Matthews S, Rao VS, Durvasula RV. 2011. Modeling horizontal gene transfer (HGT) in the gut of the Chagas disease vector Rhodnius prolixus. Parasit Vectors. 4:77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo ES, Wallau GL. 2020. Mosquito genomes are frequently invaded by transposable elements through horizontal transfer. PLoS Genet. 16:e1008946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitra R, Li X, Kapusta A, Mayhew D, Mitra RD, Feschotte C, Craig NL. 2013. Functional characterization of piggyBat from the bat Myotis lucifugus unveils an active mammalian DNA transposon. Proc Natl Acad Sci USA. 110:234–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B. 2011. How many species are there on earth and in the ocean? PLoS Biol. 9:e1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales AE, Ruedi M, Field K, Carstens BC. 2019. Diversification rates have no effect on the convergent evolution of foraging strategies in the most speciose genus of bats, Myotis. Evolution 73:2263–2280. [DOI] [PubMed] [Google Scholar]
- Moreno Santillán DD, Lama TM, Gutierrez Guerrero YT, Brown AM, Donat P, Zhao H, Rossiter SJ, Yohe LR, Potter JH, Teeling EC, et al. 2021. Large-scale genome sampling reveals unique immunity and metabolic adaptations in bats. Mol Ecol. 30:6449–6467. [DOI] [PubMed] [Google Scholar]
- Muñoz-López M, García-Pérez JL. 2010. DNA transposons: nature and applications in genomics. Curr Genomics. 11:115–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman JC, Bailey AD, Fan HY, Pavelitz T, Weiner AM. 2008. An abundant evolutionarily conserved CSB-PiggyBac fusion protein expressed in Cockayne syndrome. PLoS Genet. 4:e1000031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikaido M, Kondo S, Zhang Z, Wu J, Nishihara H, Niimura Y, Suzuki S, Touhara K, Suzuki Y, Noguchi H, et al. 2020. Comparative genomic analyses illuminate the distinct evolution of megabats within Chiroptera. DNA Res. 27:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Notwell JH, Chung T, Heavner W, Bejerano G. 2015. A family of transposable elements co-opted into developmental enhancers in the mouse neocortex. Nat Commun. 6:6644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novick P, Smith J, Ray D, Boissinot S. 2010. Independent and parallel lateral transfer of DNA transposons in tetrapod genomes. Gene 449:85–94. [DOI] [PubMed] [Google Scholar]
- Nowak RM. 1999. Walker's mammals of the world. Baltimore: (MD: ): John Hopkins University Press. [Google Scholar]
- Oliveira SG, Bao W, Martins C, Jurka J. 2012. Horizontal transfers of Mariner transposons between mammals and insects. Mob DNA. 3:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. 2011. Mobile DNA and the TE-Thrust hypothesis: supporting evidence from the primates. Mob DNA. 2:8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver KR, Greene WK. 2012. Transposable elements and viruses as factors in adaptation and evolution: an expansion and strengthening of the TE-Thrust hypothesis. Ecol Evol. 2:2912–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanski AB, Paulat NS, Korstian J, Grimshaw JR, Halsey M, Sullivan KAM, Moreno-Santillán DD, Crookshanks C, Roberts J, Garcia C, et al. 2022. Insights into mammalian TE diversity via the curation of 248 mammalian genome assemblies. bioRxiv. 2022.2012.2028.522108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JK, Feschotte C. 2007. The evolutionary history of human DNA transposons: evidence for intense activity in the primate lineage. Genome Res. 17:422–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pace JK II, Gilbert C, Clark MS, Feschotte C. 2008. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA. 105:17023–17028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagán HJ, Macas J, Novák P, McCulloch ES, Stevens RD, Ray DA. 2012. Survey sequencing reveals elevated DNA transposon activity, novel elements, and variation in repetitive landscapes among vesper bats. Genome Biol Evol. 4:575–585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagan HJ, Smith JD, Hubley RM, Ray DA. 2010. PiggyBac-ing on a primate genome: novel elements, recent activity and horizontal transfer. Genome Biol Evol. 2:293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palazzo A, Lorusso P, Miskey C, Walisko O, Gerbino A, Marobbio CMT, Ivics Z, Marsano RM. 2019. Transcriptionally promiscuous “blurry” promoters in Tc1/mariner transposons allow transcription in distantly related genomes. Mob DNA. 10:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavey CR. 2021. Comparative echolocation and foraging ecology of horseshoe bats (Rhinolophidae) and Old World leaf-nosed bats (Hipposideridae). Aust J Zool. 68:382–392. [Google Scholar]
- Peccoud J, Loiseau V, Cordaux R, Gilbert C. 2017. Massive horizontal transfer of transposable elements in insects. Proc Natl Acad Sci USA. 114:4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, Mangum SF, Ray DA. 2016. Pinpointing the vesper bat transposon revolution using the Miniopterus natalensis genome. Mob DNA. 7:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platt RN, Vandewege MW, Ray DA. 2018. Mammalian transposable elements and their impacts on genome evolution. Chromosome Res. 26:25–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritham EJ, Feschotte C. 2007. Massive amplification of rolling-circle transposons in the lineage of the bat Myotis lucifugus. Proc Natl Acad Sci USA. 104:1895–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Feschotte C, Pagan HJ, Smith JD, Pritham EJ, Arensburger P, Atkinson PW, Craig NL. 2008. Multiple waves of recent DNA transposon activity in the bat, Myotis lucifugus. Genome Res. 18:717–728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Pagan HJ, Platt RN II, Kroll AR, Schaack S, Stevens RD. 2015. Differential SINE evolution in vesper and non-vesper bats. Mob DNA. 6:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray DA, Pagan HJ, Thompson ML, Stevens RD. 2007. Bats with hATs: evidence for recent DNA transposon activity in genus Myotis. Mol Biol Evol. 24:632–639. [DOI] [PubMed] [Google Scholar]
- Reiss D, Mialdea G, Miele V, de Vienne DM, Peccoud J, Gilbert C, Duret L, Charlat S. 2019. Global survey of mobile DNA horizontal transfer in arthropods reveals Lepidoptera as a prime hotspot. PLoS Genet. 15:e1007965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhie A, McCarthy SA, Fedrigo O, Damas J, Formenti G, Koren S, Uliano-Silva M, Chow W, Fungtammasan A, Kim J, et al. 2021. Towards complete and error-free genome assemblies of all vertebrate species. Nature 592:737–746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 16:276–277. [DOI] [PubMed] [Google Scholar]
- Schaack S, Gilbert C, Feschotte C. 2010. Promiscuous DNA: horizontal transfer of transposable elements and why it matters for eukaryotic evolution. Trends Ecol Evol (Amst). 25:537–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Schwalie PC, Wilson MD, Ballester B, Gonçalves A, Kutter C, Brown GD, Marshall A, Flicek P, Odom DT. 2012. Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148:335–348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seim I, Fang X, Xiong Z, Lobanov AV, Huang Z, Ma S, Feng Y, Turanov AA, Zhu Y, Lenz TL, et al. 2013. Genome analysis reveals insights into physiology and longevity of the Brandt’s bat Myotis brandtii. Nat Commun. 4:2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva JC, Loreto EL, Clark JB. 2004. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 6:57–71. [PubMed] [Google Scholar]
- Simmons NB, Cirranello AL. Bat species of the world: a taxonomic and geographic database [Internet]. 2020. cited 04/09/2021]. Available fromhttps://batnames.org/.
- Smit AFA. 1996. The origin of interspersed repeats in the human genome. Curr Opin Genet Dev. 6:743–748. [DOI] [PubMed] [Google Scholar]
- Smit AFA, Hubley R, Green P. 2013-2015. RepeatMasker Open-4.0. Version 4.0.9.
- Sotero-Caio CG, Platt RN II, Suh A, Ray DA. 2017. Evolution and diversity of transposable elements in vertebrate genomes. Genome Biol Evol. 9:161–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffberg S, Jacobs DS, Mackie IJ, Matthee CA. 2010. Molecular phylogenetics and historical biogeography of Rhinolophus bats. Mol Phylogenet Evol. 54:1–9. [DOI] [PubMed] [Google Scholar]
- Storer JM, Hubley R, Rosen J, Smit AFA. 2021. Curation guidelines for de novo generated transposable element families. Curr Protoc. 1:e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer J, Hubley R, Rosen J, Wheeler TJ, Smit AF. 2021. The Dfam community resource of transposable element families, sequence models, and genome annotations. Mob DNA. 12:2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subudhi S, Rapin N, Misra V. 2019. Immune system modulation and viral persistence in bats: understanding viral spillover. Viruses 11:192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram V, Cheng Y, Ma Z, Li D, Xing X, Edge P, Snyder MP, Wang T. 2014. Widespread contribution of transposable elements to the innovation of gene regulatory networks. Genome Res. 24:1963–1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teeling EC, Springer MS, Madsen O, Bates P, O'Brien SJ, Murphy WJ. 2005. A molecular phylogeny for bats illuminates biogeography and the fossil record. Science 307:580–584. [DOI] [PubMed] [Google Scholar]
- Teeling EC, Vernes SC, Dávalos LM, Ray DA, Gilbert MTP, Myers E. 2018. Bat biology, genomes, and the Bat1K project: to generate chromosome-level genomes for all living bat species. Annu Rev Anim Biosci. 6:23–46. [DOI] [PubMed] [Google Scholar]
- Thomas J, Phillips CD, Baker RJ, Pritham EJ. 2014. Rolling-circle transposons catalyze genomic innovation in a mammalian lineage. Genome Biol Evol. 6:2595–2610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Schaack S, Pritham EJ. 2010. Pervasive horizontal transfer of rolling-circle transposons among animals. Genome Biol Evol. 2:656–664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas J, Sorourian M, Ray D, Baker RJ, Pritham EJ. 2011. The limited distribution of Helitrons to vesper bats supports horizontal transfer. Gene 474:52–58. [DOI] [PubMed] [Google Scholar]
- Threlfall J, Blaxter M. 2021. Launching the Tree of Life Gateway. Wellcome Open Research 6:125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tipney HJ, Hinsley TA, Brass A, Metcalfe K, Donnai D, Tassabehji M. 2004. Isolation and characterisation of GTF2IRD2, a novel fusion gene and member of the TFII-I family of transcription factors, deleted in Williams–Beuren syndrome. Eur J Hum Genet. 12:551–560. [DOI] [PubMed] [Google Scholar]
- Trizzino M, Park Y, Holsbach-Beltrame M, Aracena K, Mika K, Caliskan M, Perry GH, Lynch VJ, Brown CD. 2017. Transposable elements are the primary source of novelty in primate gene regulation. Genome Res. 27:1623–1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallau GL, Ortiz MF, Loreto ELS. 2012. Horizontal transposon transfer in eukarya: detection, bias, and perspectives. Genome Biol Evol. 4:801–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Tian S, Galindo-González J, Dávalos LM, Zhang Y, Zhao H. 2020. Molecular adaptation and convergent evolution of frugivory in Old World and neotropical fruit bats. Mol Ecol. 29:4366–4381. [DOI] [PubMed] [Google Scholar]
- Wang T, Zeng J, Lowe CB, Sellers RG, Salama SR, Yang M, Burgess SM, Brachmann RK, Haussler D. 2007. Species-specific endogenous retroviruses shape the transcriptional network of the human tumor suppressor protein p53. Proc Natl Acad Sci USA. 104:18613–18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells JN, Feschotte C. 2020. A field guide to eukaryotic transposable elements. Annu Rev Genet. 54:539–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O, et al. 2007. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 8:973–982. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Cheng TC, Huang G, Lu Q, Surleac MD, Mandell JD, Pontarotti P, Petrescu AJ, Xu A, Xiong Y, et al. 2019. Transposon molecular domestication and the evolution of the RAG recombinase. Nature 569:79–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G, Cowled C, Shi Z, Huang Z, Bishop-Lilly KA, Fang X, Wynne JW, Xiong Z, Baker ML, Zhao W, et al. 2013. Comparative analysis of bat genomes provides insight into the evolution of flight and immunity. Science. 339:456–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H-H, Peccoud J, Xu M-R-X, Zhang X-G, Gilbert C. 2020. Horizontal transfer and evolution of transposable elements in vertebrates. Nat Commun. 11:1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuo X, Rho M, Feschotte C. 2013. Genome-wide characterization of endogenous retroviruses in the bat Myotis lucifugus reveals recent and diverse infections. J Virol. 87:8493–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All assemblies are available on GenBank (see supplementary table S1 , Supplementary material online for accession numbers). TE consensus sequences, and their seed alignments, are available via the Dfam database. All other data is available in the supplementary materials, Supplementary Material online; code used in the analysis is available at github.com/daray/bat_ht.
Conflict of interest statement. The authors declare no potential conflicts of interest with respect to the authorship and/or publication of this article.