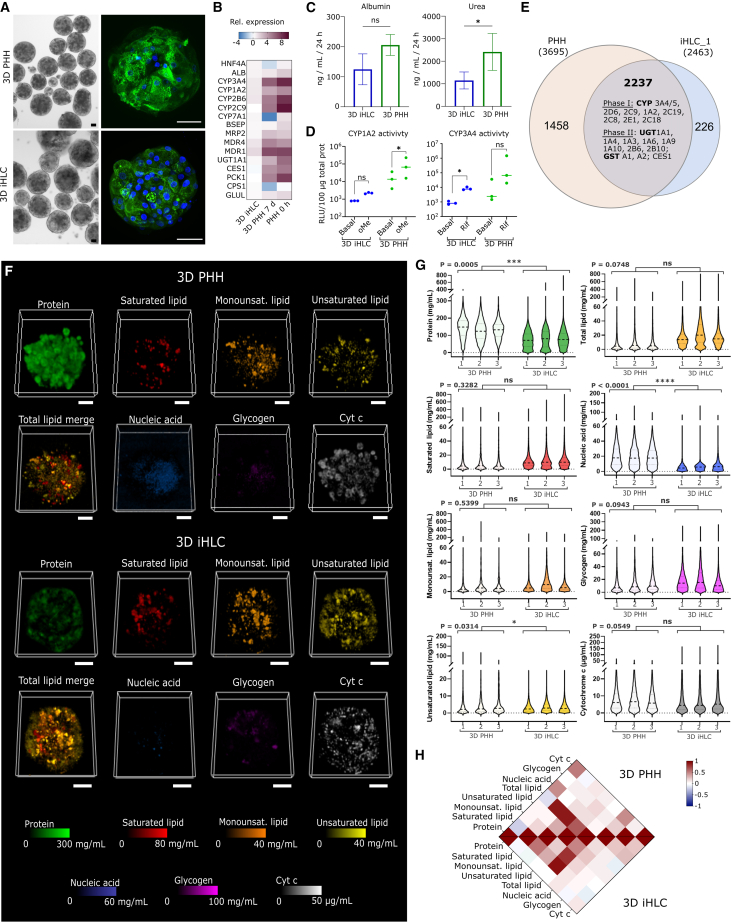
Figure 2.
qRamanomics enables quantitative high-content composition comparison between primary human hepatocyte spheroids and induced hepatocyte-like cell organoids
(A) Representative bright-field and immunofluorescence images of 3D PHHs and 3D iHLCs (green, albumin; blue, nuclei). Scale bar, 50 μm.
(B) Heatmap shows mean relative expression of selected hepatocyte-specific genes given as log2. iHLC organoids from iPSC line WTSIi028-A (iHLC_3) were used for the normalization.
(C) Secretion of albumin and urea in PHH and iHLC organoids. (p = 0.059 for albumin secretion, p = 0.0386 for urea secretion.)
(D) Basal and induced cytochrome P450 activity. Data represent mean for three donors of 3D PHHs and three cell lines of 3D iHLCs. (CYP1A2 activity in iHLC, p = 0.057, in PHH p = 0.039; CYP3A4 activity in iHLC, p = 0.0266, in PHH p = 0.3902.)
(E) Venn diagram of proteins detected in PHH (n = 1 donor) and iHLC organoids (n = 1 cell line).
(F) High-content quantitative Ramanomic imaging reveals distribution of molecular content throughout 3D PHH spheroids from a single donor (n = 3) and 3D iHLC organoids (n = 3). Scale bars, 50 μm.
(G) Inter-spheroid/organoid repeatability within and between specimen groups. Nested t test revealed significant differences in protein, unsaturated lipid, and nucleic acid content between 3D PHH and 3D iHLC samples. (p values are indicated on the figure itself.)
(H) Pearson’s correlation chemometric heatmaps illustrate co-localization of various molecular components in 3D PHH and iHLC.