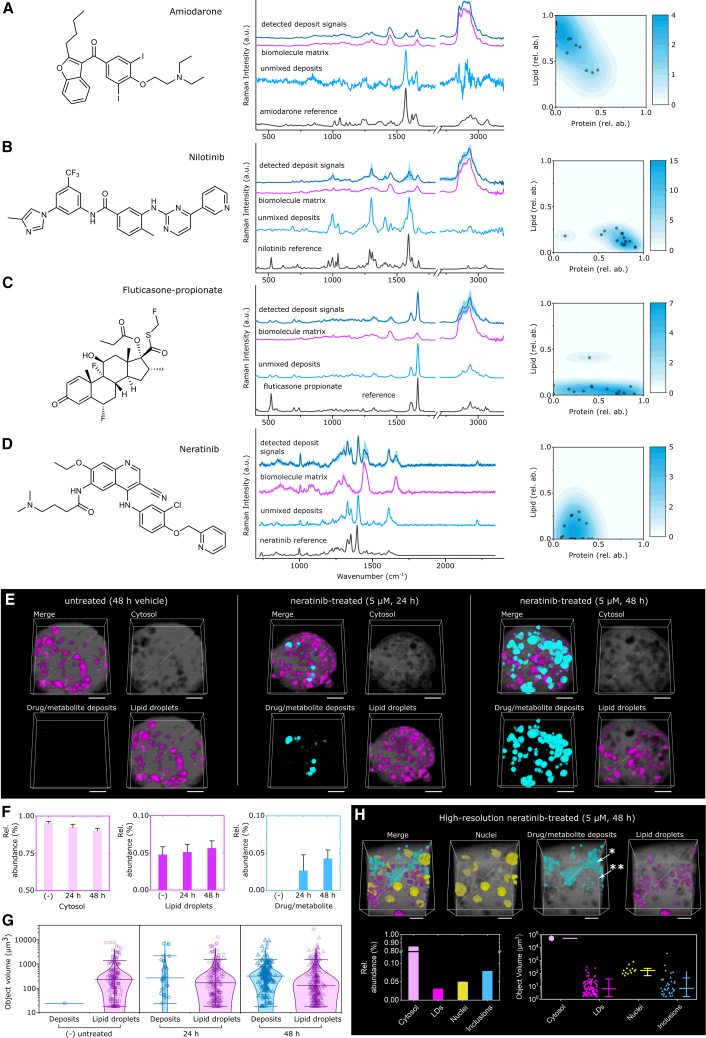
Figure 4.
Drug and drug metabolite deposits were detected within lysosomes yielding altered Raman spectral signals as compared with parent drug for drug-treated 3D iHLC organoids
(A–D) Chemical formulas for drugs whereby detected deposit signals were extracted from intracellular drug and/or metabolite aggregate regions of interest (five deposits from n = 3 3D iHLCs for each treatment group) for (A) amiodarone, (B) nilotinib, (C) fluticasone propionate, and (D) neratinib-treated 3D iHLC organoids. Spectral difference between unmixed deposits and parent drug reference signals suggests metabolism of compounds.
(E and F) (E) qVRI reveals distribution of drug/metabolite (cyan) and lipid droplets (magenta) for neratinib-treated 3D PHHs (scale bars, 50 μm). (F) Relative volumetric abundance for each component.
(G) Particle size distribution analysis of drug/metabolite deposits (inclusions) within drug-treated and control specimens (n = 3 organoids per group).
(H) High-resolution 3D Raman chemical imaging suggests neratinib accumulation in lysosomes (∗∗) and excretion to bile canaliculi (∗). The mean and error bars are plotted adjacent to the data points to aid clarity. Scale bars, 10 μm.