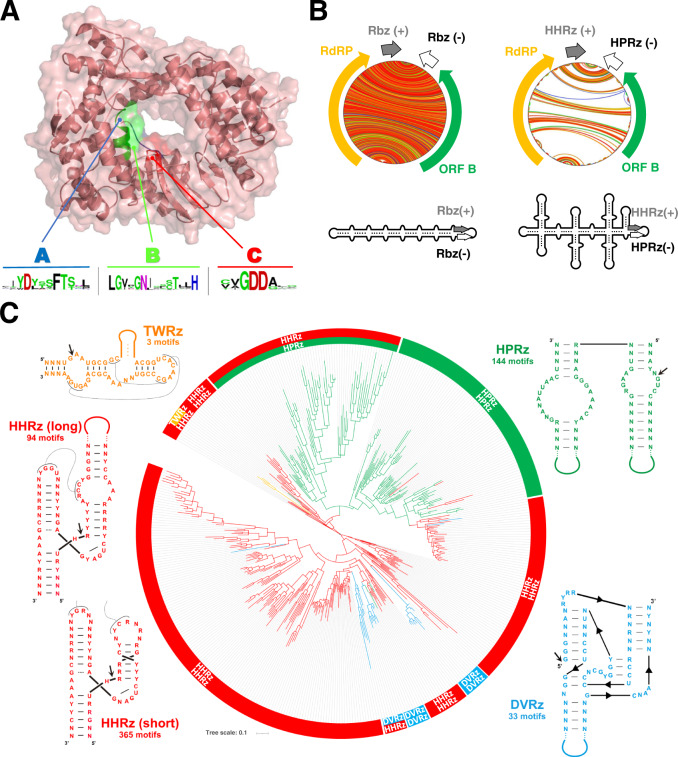
Fig. 2. Ambivirus RNA-dependent RNA polymerase phylogeny.
A ColabFold-AlphaFold2 predicted structure of the representative Armillaria borealis ambivirus 2 (Accession MW423810.1) ORF-A shows a classic polymerase palm fold structure. Conservation of catalytic residues in motif A, B, and C is seen across all ambiviruses identified in this study and present in databases. B Circular plots of the predicted RNA secondary structures of two representative ambiviruses. On the left, an example of the characteristic rod-like structure predicted for most ambiviruses. On the right, an example of a highly branched architecture predicted for some ambivirus RNA genomes, usually carrying a mixed pair of HHRz/HPRz motifs. C Maximum-likelihood phylogenetic tree of the RNA-dependent RNA polymerase palmprints from the 439 distinct species-like operational taxonomic units (sOTUs). A more detailed view of the tree can be found at the Supplementary Fig. 7. There are up to six major clades of ambiviruses showing diverse ribozyme usage. Up to 60% sOTUs use paired HHRz very similar to the motifs from Epsilonviruses14,41 or plant viroids4, whereas ~30% carry either two HPRz or a HHRz/HPRz mix, 5% carry the DVRz (either paired or a mix with HHRz) characteristic of animal deltaviruses, and three sOTUs contain a mix of the TWRz (P1 architecture)42 and the HHRz motifs. Consensus ribozyme structures (weighted nucleotide conservation threshold of 70%) of twister (TWRz, in orange), the hammerhead (HHRz, in red, two length variants), hairpin (HPRz, in green) and Delta virus (DVRz, in blue) ribozymes present in ambiviruses are shown. Site of self-cleavage is indicated with an arrow.