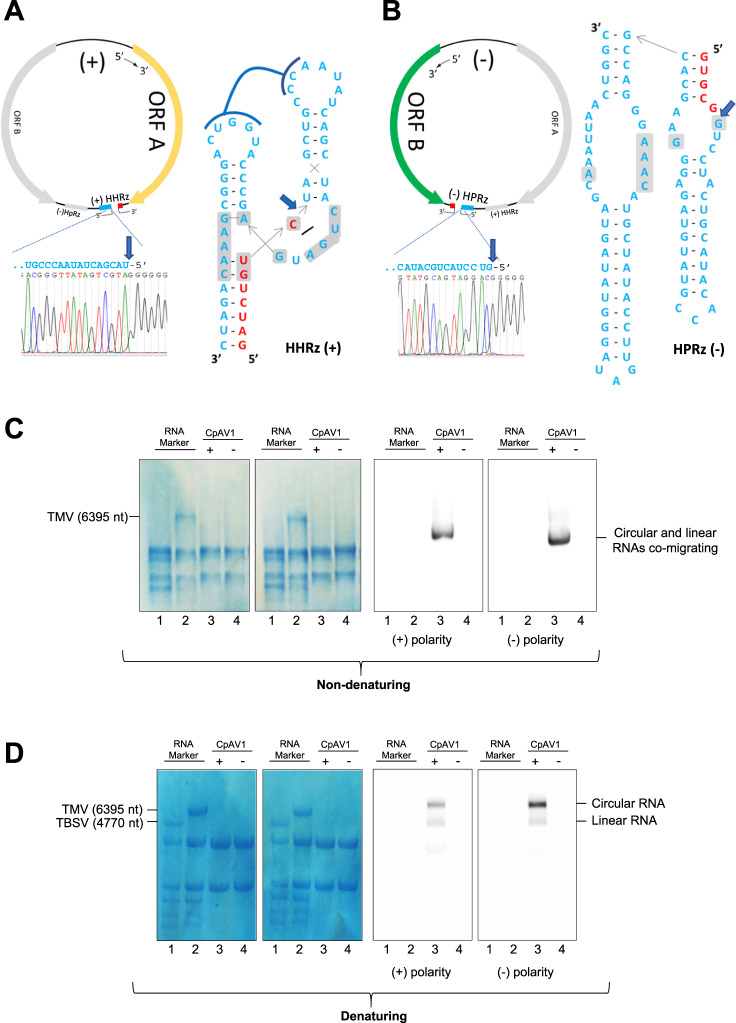
Fig. 3. Ambiviruses encode functional +/− strand ribozymes and have circRNA genomes.
5’ RACE and Sanger sequencing of the (A) positive and (B) negative strand of TuAmV4 genomic RNAs show in vivo self-cleavage (blue arrow) at the predicted cut-sites for its HHRz and HPRz motifs, respectively. Ribozyme secondary structures drawn as inlay. C, D Northern blot assays of RNA preparations separated by electrophoresis under non denaturing (C) and denaturing (D) conditions and hybridized with probes specific for the (+) and (−) CpAV1 RNA; Lanes 3 and 4 are RNAs from CpAV1-infected and non-infected Cryphonectria parasitica, respectively; RNA markers are RNAs from plants, healthy or TBSV-infected (line 1 in C and D, respectively) and TMV-infected (line 2 in C and D). The single signal detected in the infected isolate with the either probe under non-denaturing conditions corresponds to both circular and linear CpAV1 genomic RNAs (4663 nt), which are expected to co-migrate (C). In denaturing conditions (D), the ambivirus circRNAs are delayed with respect to the respective linear forms (lower band). In C, D, the two leftmost panels are total RNA stained with methylene blue. The same results were obtained from three independent experiments.