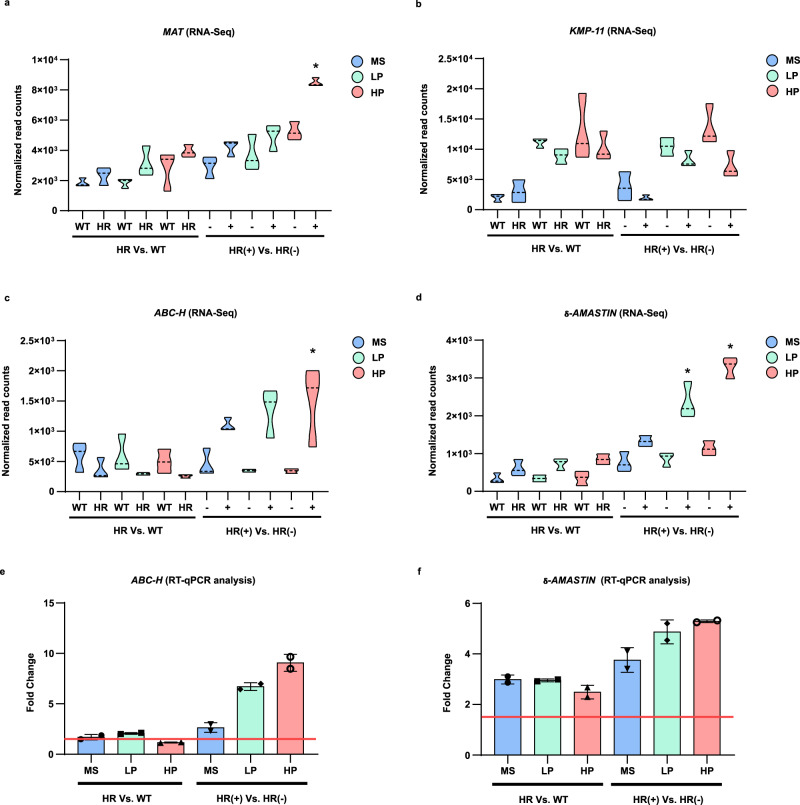
Fig. 5. Transcripts associated with the mechanisms of SbIII drug resistance show a shift in translational efficiency.
RNA-Seq (normalized read count) and RT-qPCR (mRNA fold change) were used to represent the changes in translational efficiency per polysome fraction and gene target. The Y axis represents the read counts normalized by DESeq2’s median of ratios. The X axis shows polysome fraction distributed per experimental condition in two panels, basal changes (left panel in each graph) and changes to mediate drug resistance (right panel in each graph). Drug-treated (+), not treated (–). a The S-adenosylmethionine synthetase (MAT, LmjF.30.3500) typically upregulated in SbIII-resistant parasites. P value (MS): 0.84, P value (LP): 0.11, P value (HP): 0.009 (*). b The Kinetoplastid membrane protein-11 (KMP-11, LmjF.35.2210) is typically downregulated in SbIII-resistant parasites. P value (MS): 0.76, P value (LP): 0.50, P value (HP): 0.094. c The ATP-binding cassette protein subfamily H (ABC-H, LmjF.11.0040) is typically upregulated in SbIII-resistant parasites. P value (MS): 0.69, P value (LP): 0.06, P value (HP): 1.33 × 10−05(*). d δ-amastin (LmjF.31.0450) a surface protein with an unknown role in SbIII-resistance. P value (MS): 0.63. P value (LP): 0.02 (*). P value (HP): 7.53 × 10−10 (*). a–d Two-sided Wald test. Multiple comparison corrected by the Benjamini–Hochberg Procedure. Adjusted P value for drug challenge condition (right side of the figure). n: three biologically independent replicates. The shift in translational efficiency was also detected by RT-qPCR analysis. e Expression of ABC-H as linear fold change distributed per polysome fraction and experimental condition after qPCR analysis. f Expression of δ-amastin as linear fold change distributed per polysome fraction and experimental condition after RT-qPCR analysis. e, f n: two independent biological replicates with three technical replicates. Data are presented as mean values +/– SD. Threshold of significant fold change ≥1.5 (horizontal red line) for RT-qPCR. Monosomes (MS), light polysomes (LP), and heavy polysomes (HP). The raw data of (a–f) are available in Source Data.