Fig. 1.
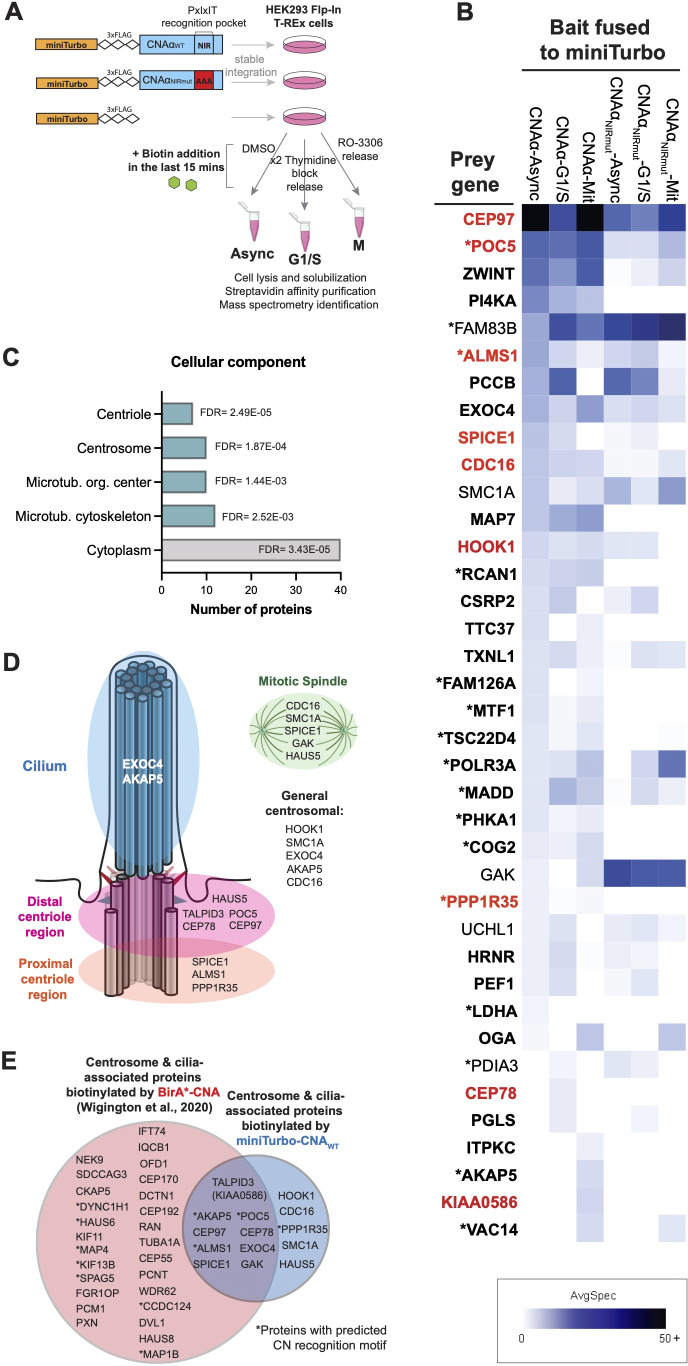
Calcineurin is proximal to centrosome proteins. (A) Scheme for cell cycle synchronization and PDB–MS. Amino acid residues mutated in the PxIxIT recognition pocket are indicted. Async, asynchronous; M, mitosis. (B) Heat map showing average spectral counts (AvgSpec) of the CN-proximal proteins (from DDA mass spectrometry) in each of the indicated experimental conditions. Proteins with PxIxIT docking-dependent biotinylation [log2 spectral count ratio (CNAαWT/CNAαNIRmut)≥0.5] are shown in bold type. Proteins annotated by the GO term ‘centrosome’ are shown in red. Asterisks indicate proteins containing PxIxIT or LxVP motifs. Mit, mitosis. (C) Cellular component GO terms with statistically significant enrichment among the CN-proximal proteins. The number of CN-proximal proteins assigned to each term is plotted, and the FDR is indicated. (D) CN-proximal proteins with previously reported localization to centrosomes, cilia or mitotic spindles. See Table S2 for references. (E) Overlap of centrosome and cilia CN-proximal proteins from Wigington et al. (2020) (pink circle) and this study (blue).