Graphical Abstract
Graphical Abstract.
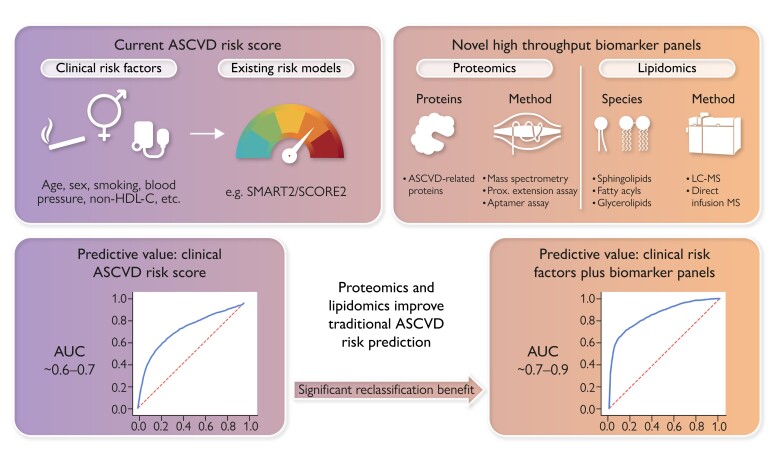
Proteomics and lipidomics improve traditional ASCVD risk prediction. Plasma proteomics and lipidomics hold a major promise in improving ASCVD risk prediction offering high-throughput assessment using different techniques. Albeit retrospectively, large proteomic and lipidomic studies have consistently demonstrated improved ASCVD risk prediction in terms of discrimination and reclassification benefit compared to risk scoring with clinical characteristics. Future studies into clinical utility are needed for widespread clinical implementation. ASCVD, atherosclerotic cardiovascular disease; AUC, area under the receiver operating curve; non-HDL-C, non-high-density lipoprotein cholesterol; SCORE2, Systematic COronary Risk Evaluation 2 system; SMART2, Second Manifestations of ARTerial disease 2; LC-MS, liquid chromatography–mass spectrometry; MS, mass spectrometry.
Keywords: Proteomics, Lipidomics, Multiomics, ASCVD, Risk score
Abstract
Given the limited accuracy of clinically used risk scores such as the Systematic COronary Risk Evaluation 2 system and the Second Manifestations of ARTerial disease 2 risk scores, novel risk algorithms determining an individual’s susceptibility of future incident or recurrent atherosclerotic cardiovascular disease (ASCVD) risk are urgently needed. Due to major improvements in assay techniques, multimarker proteomic and lipidomic panels hold the promise to be reliably assessed in a high-throughput routine. Novel machine learning-based approaches have facilitated the use of this high-dimensional data resulting from these analyses for ASCVD risk prediction. More than a dozen of large-scale retrospective studies using different sets of biomarkers and different statistical methods have consistently demonstrated the additive prognostic value of these panels over traditionally used clinical risk scores. Prospective studies are needed to determine the clinical utility of a biomarker panel in clinical ASCVD risk stratification. When combined with the genetic predisposition captured with polygenic risk scores and the actual ASCVD phenotype observed with coronary artery imaging, proteomics and lipidomics can advance understanding of the complex multifactorial causes underlying an individual’s ASCVD risk.
Introduction
Identification of patients at greatest risk of atherosclerotic cardiovascular disease (ASCVD) events poses a major challenge in both primary and secondary prevention.1–3 The vast majority of primary prevention patients at greatest risk remains unidentified prior to their first clinical event.4 In secondary prevention, the ASCVD event recurrence rate remains high despite adhering to guideline recommendations for treatment.5 To effectively reduce the increasing global burden of ASCVD, improvement in risk stratification is of essence. Currently used clinical risk algorithms, including the Framingham risk score, the Systematic COronary Risk Evaluation 2 (SCORE2) system, and the Second Manifestations of ARTerial disease 2 (SMART2),6–10 are based on traditional risk factors for cardiovascular disease and, retrospectively, predict future events with limited accuracy. The current clinical risk scores include risk factors such as smoking, hypertension, diabetes, and hypercholesterolaemia that cannot encompass the multitude of pathophysiological processes contributing to the onset and progression of ASCVD, nor do these scores incorporate the heterogeneity in interindividual atherogenic vulnerability. The limitations of current risk prediction algorithms are an even more urgent problem in view of the rapidly expanding armamentarium of ASCVD risk lowering medications such as inclisiran,11,12 sodium–glucose cotransporter 2 inhibitors,13–15 glucagon-like peptide-1 agonists,16,17 bempedoic acid,18,19 and anti-inflammatory agents,20,21 which due to their high costs mandate to restrict use in patients at highest risk or patients who are likely to benefit the most from the given therapy.22 Thus, there is an urgent need for reliable instruments determining an individual’s susceptibility of future or recurrent ASCVD risk.
Over the last years, we have witnessed several promising developments which may prove useful to optimize a more personalized risk approach. Such developments include genetic tools such as the use of polygenic risk scores, which allow the identification of genetic predisposition (or ‘vulnerability’) towards ASCVD risk and are in part independent from the traditional ASCVD risk factors.23–25 In parallel, technical improvements in imaging modalities, comprising coronary artery calcium scoring and coronary computed tomography angiography (CCTA), have facilitated a further improvement in individualized risk prediction with good reproducibility especially when analyzed using artificial intelligence.26,27 Apart from genetic markers and capturing structural changes in the arterial wall by imaging, many efforts have focused on the incremental value of blood biomarkers as a ‘liquid biopsy’ of either causal or sequential markers for ASCVD risk. At present, few biomarkers are proposed for use in ASCVD risk prediction, i.e. high-sensitivity C-reactive protein.28 Notably, plasma markers such as N-terminal pro-brain natriuretic peptide (NT-proBNP) and high-sensitivity troponin, when used in isolation, have a relatively limited additive value over traditional risk factors including age in ASCVD risk prediction.29,30 Thus, attention has shifted towards the use of ‘-omics’ approaches, in particular proteomics and lipidomics. These two major molecular moieties, proteins and lipids, have both shown to hold promise to further improve ASCVD risk prediction.31–39
Plasma proteomics is a rapidly evolving field of research. The term ‘proteome’ was first introduced by Wilkins and Williams in 1994.40 The plasma proteome refers to all proteins present in plasma.41 The plasma proteome is the most complex proteome in the human body. The concentrations of proteins in plasma span over 12 orders of magnitude in linear dynamic range. In the past few years, development and optimization of protein detection assays have enabled the measurement of thousands of plasma proteins.
In addition to the plasma proteome, the plasma lipidome has attracted interest in the omics revolution for ASCVD risk prediction thanks to rapid technological advancements in mass spectrometry (MS). The numerous plasma lipid species can be categorized in six major lipid classes: fatty acyls, glycerophospholipids, glycerolipids, sphingolipids, sterols, and prenols.42 Of these categories, sphingolipids (ceramide species), fatty acyls, and glycerolipids (triglyceride species) have attracted most attention in clinical studies.43
Despite these technological advances and encouraging first results over the last years, neither multimarker proteomics nor lipidomics panels have been adopted by current prevention guidelines.6 Thus, this review focuses on a key question: can targeted plasma proteomics and lipidomics meaningfully improve ASCVD risk prediction?
The discovery pipeline towards targeted proteomics or lipidomics panels
The process of identifying biomarker candidates with prognostic value for ASCVD events should start with an untargeted discovery approach. This approach can identify novel lipids and proteins potentially relating to ASCVD risk. After successful identification of ASCVD-related biomarker candidates, a targeted approach can be used to quantify levels of previously identified biomarker levels in a high-throughput routine, e.g. to improve ASCVD risk stratification. For lipidomics, the targeted approaches generally rely on similar (MS-based) techniques compared to the discovery approach, while targeted proteomics analyses generally use different methods for high-throughput use.
Analytical approaches for discovery proteomics
The use of discovery proteomics enables the detection of proteins in a particular sample without a priori selection of proteins. For such an analysis, MS is mainly used (although large panel assays with preselected proteins such as SomaScan or Olink have also been reported for discovery). Plasma proteins are subjected to enzymatic digestion giving rise to peptides (bottom-up approach).44 The mass spectrometer analyses these peptides and the resulting spectra can be matched to specific proteins using a database or a spectral library. This unbiased approach allows for identification of thousands of proteins without the need for binders. Importantly, discovery proteomics using MS is biased towards the more abundant plasma proteins. This lack of sensitivity of MS in the low abundant protein range can be circumvented using several techniques such as depletion of high-abundance proteins, prefractionation, or nanoparticle enrichment.45,46
Analytical approaches for targeted proteomics
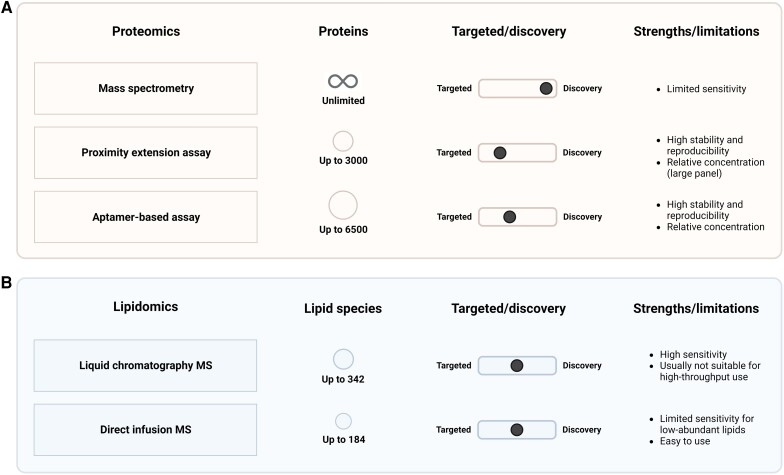
For high-throughput quantification of a set of particular proteins, a targeted proteomics approach can be used. Technological advances over the last years have resulted in multiple methods to clinically perform such targeted proteomics. Using selective reaction monitoring, multiple reaction monitoring, or parallel reaction monitoring, MS can be used to measure specific peptides. However, the targeted MS approaches also fail to detect very low abundant plasma proteins without sample preparation, similar to the discovery of MS.47 Apart from MS, the most widely used methods for targeted proteomics are classical immunoassays, proximity ligation assays, and aptamer-based assays. Such approaches—although measuring only a predetermined subset of proteins—allow for high-throughput assessment of a large number of proteins. Over the last years, two techniques have been the most widely used: proximity extension assays (Olink Biosciences, Uppsala, Sweden) and aptamer technology (SomaScan Assay, Somalogic, Boulder CO, USA) (Figure 1A). For every protein, the proximity extension assays use an antibody pair labelled with unique DNA oligonucleotides, thereby reducing cross-reactivity and increasing specificity.48 The most recent version of the assay is able to measure 3072 different proteins in plasma. Once both antibodies have bound their specific epitopes on the protein, the DNA oligonucleotides hybridize and are extended after addition of DNA polymerase. Using quantitative polymerase chain reactions (qPCR) or next-generation sequencing as a readout, a relative concentration is returned for every protein on the panel. The Olink panels have shown good reproducibility and stability for the vast majority of proteins on the panels.49 In contrast, the SomaScan assay uses a protein affinity-based approach with modified aptamers rather than antibodies to quantify relative protein concentrations in plasma.50,51 These modified aptamers are single-stranded DNA (or RNA) oligonucleotides which can bind to specific proteins in a highly multiplex platform. The most recent assay can measure up to 6596 unique proteins.49 Importantly, both the large proximity extension assay panels and the aptamer-based assay result in relative protein values rather than absolute protein concentrations. Of note, however, the most recent smaller panels with the proximity extension assay from Olink (48 proteins) also allow for absolute protein quantification.
Figure 1.
Methods for performing plasma proteomics and lipidomics. Characteristics of the most frequently used analysing methods for plasma proteomics (A) and plasma lipidomics (B) in large-scale studies. Shown are differences in proteins/lipid species, target/discovery, and strengths/limitations. The numbers shown reflect the maximum number of proteins/lipid species of the used techniques in the clinical studies (Tables 1 and 2) included in the current review. Created with BioRender.com. MS, mass spectrometry.
Two recent studies have compared the targeted proteomics approaches by Olink and Somalogic.52,53 Approximately one-third of ∼600 overlapping proteins from both platforms showed at best a modest correlation between platforms, and another one-third only showed medium correlations.53 Thus, there are discrepancies between these two different platforms, possibly caused by unspecific binding, protein interactions, post-translational modifications, protein truncation, and proteolytic cleavage products, which are likely not to be identified.54 Overall, the Olink platform showed more intra- and interassay variability, but was more accurate when comparing the platform protein levels to genetic readouts and protein quantification by conventional enzyme-linked immunosorbent assays (ELISA), compared to the Somalogic platform.53 A previous study showed that SomaScan—since the modified aptamers mostly rely on a single binder (in contrast to the proximity extension assay with an antibody pair)—has limited specificity for certain proteins, e.g. growth differentiation factor (GDF)-8 and GDF-11.55 Although both panels allow for high-throughput protein profiling, these limitations in specificity and reproducibility warrant further investigations and must be overcome before clinical implementation can be realized.
Next to these high-throughput and large panel approaches, there are several other non-MS proteomic technologies with smaller panels available, such as the Luminex, MesoScale Discovery, and ProterixBio.56 Limited sensitivity and cross-reactivity observed with these assays have restricted their application in large, high-throughput settings.57,58 Chip-based proteomic technologies currently in development hold promise and could advance the current biomarker field.59 Finally, a novel MS-based approach, data-independent acquisition, combines both discovery and targeted proteomics approaches.60 This technique enables the combination of a broad coverage spectrum with the high-throughput properties of a targeted proteomics approach, however is still limited by a lack of sensitivity.61
Analytical approaches for lipidomics
For lipidomics, methods for both discovery and targeted lipidomics rely on MS. Mass spectrometry can be coupled to prior separation by gas chromatography (GC-MS) or liquid chromatography (LC-MS). Alternatively, lipid extracts can be directly infused (direct-infusion MS). The latter two approaches are the most widely used (Figure 1B). Liquid chromatography–mass spectrometry first separates the lipid species based on their chemical properties by liquid chromatography before ionization and detection by MS, whereas direct-infusion MS infuses lipid species without prior separation before MS.62,63 The advantage of LC-MS is that it is a very accurate and sensitive method because of the separation of lipid species by LC, while direct-infusion MS is less sensitive for low-abundant lipid species but is easier to use and can capture many lipid species at once.43 One of the most widely used lipidomics platforms, developed as well as clinically validated by Zora Diagnostics, measures the levels of four ceramide species in plasma using LS-MS.64 These ceramide levels are usually interpreted in ratios—thereby reducing impact of sample variability—which reduces intra- and interassay variability.64 Other platforms that have been used in ASCVD studies were mainly LC-MS-based techniques detecting hundreds of lipid species.
Atherosclerotic cardiovascular disease risk prediction with high-dimensional proteomics or lipidomics data
The vast majority of proteomics or lipidomics analyses results in high-dimensional data sets including up to 1000 variables. This contrasts large studies into ASCVD risk prediction with traditional clinical characteristics where the number of patients greatly exceeds the number of predictors (P<<n). In most proteomics or lipidomics studies, the amount of proteins or lipid species analyzed is comparable to the number of patients (P≈n). However, with the assay improvements over the last years, novel studies will have to deal with more predictors than study subjects (P > n). This abundance of variables usually means that traditional regression modelling can no longer be used for ASCVD risk prediction if one considers the traditionally used rule of thumb of ‘one variable in 10 subjects’.65 To analyse these high-dimensional data sets without overfitting, a relatively novel but rapidly improved approach is needed: machine learning.
There are several statistical and machine learning approaches which have been used in large-scale biomarker studies for ASCVD risk prediction deserving closer attention: least absolute shrinkage and selection operator (LASSO) for traditional regression models, artificial neural networks, and extreme gradient boosting (XGBoost). Least absolute shrinkage and selection operator is one of the oldest methods used and can be used for variable selection and regularization for traditional regression models.66,67 The L1 regularization in the LASSO regression will result in shrinkage of coefficients to zero and therefore can lead to elimination of variables from the model (dimensionality reduction).61 While relatively simple to use, a limitation of the traditional regression method is that it cannot account for non-linear relationships and between-variable interactions which are usually present in large-scale biomarker studies.61 For these purposes, more sophisticated machine learning-based approaches such as artificial neural networks or gradient boosting can be applied. However, these machine learning models still need to be coupled with dimensionality reduction techniques to avoid risk of overfitting.68–70 Machine learning methods are especially useful in studies with numerous biomarkers (e.g. >50) since they can combine many biomarkers modeling their complex interactions which are not detected with traditional statistical approaches. Artificial neural networks were created to mimic biological neural networks.71 In general, they are very efficient in difficult tasks (e.g. image recognition) but are complex to understand and need a large amount of data. Extreme gradient boosting combines an ensemble of weak prediction trees and performs well in generalization between studies thanks to arbitrary loss functions.72 In general, gradient boosting machines are easy to use and perform well in accuracy and external validation of a model. A major advantage of the artificial neural networks as well as the gradient boosting machines over traditional statistical modelling is that they can cope with a larger number of variables, non-linear relationships, and between-variable interactions.
Measuring model performance in atherosclerotic cardiovascular disease risk prediction
After training a prediction model in a particular data set, the goal is to validate its performance in predicting a particular outcome. Especially with machine learning models—which have more flexibility than traditional regression models—there is a significant risk of overfitting the developed model on the training data set.73 Overfitting implies that the model is trained (too) well with the data set correlations whereas it may perform poor in other populations or data sets, thus limiting its generalizability. Therefore, it is of utmost importance to validate a developed model on data that was not used to train the model. This can be achieved using internal validation: splitting the initial data set into a training set and a separate test set to measure model performance. This can be repeated multiple times, using different portions of the dataset (k-fold cross-validation) to train and test the model. The optimal method of model validation is external validation: validating the constructed model in an independent data set or cohort. In well-conducted machine learning studies, one can compare the performance in the derivation cohort to the performance in the independent validation cohort to test the generalizability of the developed model.
To evaluate model performance, there are three major types of performance measures with clinical value: discrimination, calibration, and reclassification.
Discrimination refers to the ability of the model to discriminate between subjects with and without the outcome. It is used to measure classification performance on a binary outcome, e.g. the occurrence of an ASCVD event. The area under the receiver operating curve (AUC) or C-statistic (which are identical for a binary outcome) is mostly used to assess the discrimination performance.74 These receiver operating characteristic (ROC) curve plots illustrate the sensitivity and the specificity of the test for different cut-offs chosen.
Calibration is an important measure to assess the clinical performance of a model. It specifies the relationship between the predicted probability of a certain outcome and the actual observed frequency of the outcome. In other words, will the predicted risk in a certain risk group correspond to the observed risk in this group? For a binary outcome (event/non-event), calibration can be displayed using a calibration plot with the predicted probability on the x-axis and the observed event rate on the y-axis. This can be done using decile or quintile groups of predicted probabilities. If perfectly calibrated, the intercept of the calibration plot will be exactly 0 while the slope of the calibration plot will be 1.
Reclassification relates to the ability of a novel model to correctly reclassify subjects compared to a reference model. It can be used to estimate the additive value of the new model compared to the clinical standard or existing model. Upward improvement of subjects with the outcome compared to the reference model will improve classification, while downward reclassification in subjects with the outcome reduces classification accuracy. This reclassification can be presented in a reclassification table or with reclassification measures such as the net reclassification improvement (NRI) or the integrated discrimination improvement (IDI).74,75 Due to the different methods used for the calculation of reclassification indices, particularly for the NRI (e.g. categorical and continuous),76 these indices should be interpreted with caution and cannot be compared directly between most studies.
Recent and key studies using proteomics for atherosclerotic cardiovascular disease risk prediction
The first studies into ASCVD risk prediction with relative small targeted plasma proteomics panels were performed in the Framingham Heart Study, published approximately 10 years ago.77,78 Since then, targeted plasma proteomics signatures have been correlated to a plethora of disease phenotypes, such as coronary plaque morphology,79 stroke,80 and type 2 diabetes mellitus.81 Several studies during the last years have focused on developing and validating a protein risk model for clinical prediction of ASCVD events (Table 1).
Table 1.
Clinical key studies using proteomics for ASCVD risk prediction
| Study and year | Study population | Protein platform | Method of analysis | Outcome | Validation performance compared to clinical model | |
|---|---|---|---|---|---|---|
| Ganz et al., 201633 | Derivation: stable CHD, 938 subjects (Heart and Soul study) | Modified aptamers (SomaScan), 1130 proteins | LASSO variable selection, Cox regression prediction model | Composite: MI, stroke, TIA, HF hospitalization, all-cause mortality | 9-protein model: | |
| AUC: 0.70 (95% CI 0.67–0.72; ΔAUC 0.05) | ||||||
| Validation: population-based study, stable CHD, 971 subjects (HUNT3) | NRI: 0.43 (95% CI 0.26–0.57) | |||||
| IDI: 0.08 (95% CI 0.05–0.10) | ||||||
| Ho et al., 201878 | 3523 subjects without ASCVD (Framingham Heart Study), internal validation | Modified ELISA (Luminex), 85 proteins | Cox regression | Composite: non-fatal MI, PCI, CABG, ischaemic stroke, CHD death | 1-protein (GDF-15) added to clinical model: | |
| AUC: 0.76 (ΔAUC 0.01) | ||||||
| NRI (2 category): 0.01 (95% CI -0.02–0.07) | ||||||
| IDI: 0.01 (95% CI 0.00–0.02) | ||||||
| Hoogeveen et al., 202031 | Derivation: primary prevention case–control, 822 subjects (EPIC-Norfolk) | Proximity extension assay (Olink), 368 proteins | Extreme gradient boosting machine learning with stability selection | MI (derivation); composite: subclinical atherosclerosis and ASCVD events (validation) | 50-protein model: | |
| AUC: 0.71 ± 0.07 (ΔAUC 0.10) | ||||||
| Validation: primary prevention case-control, 702 subjects (PLIC) | ||||||
| NRI: 0.07 (short-term prediction) | ||||||
| Unterhuber et al., 202135 | Derivation: 1998 subjects at increased ASCVD risk (LIFE-Heart Study) | Proximity extension assay (Olink), 92 proteins | Logistic regression, Cox regression, extreme gradient boosting, neural networks | All-cause mortality | 92-protein model (extreme gradient boosting, binary outcome): | |
| Validation: primary prevention, 772 subjects (PLIC) | AUC: 0.91 (95% CI 0.86–0.95; ΔAUC 0.24) | |||||
| NRI: 0.16 (compared to logistic regression) | ||||||
| Nurmohamed et al., 202232 | Derivation: secondary prevention, 870 subjects (SMART) | Proximity extension assay (Olink), 276 proteins | Extreme gradient boosting machine learning with stability selection | Composite: acute MI, ischaemic stroke, cardiovascular death | 50-protein model: | |
| AUC: 0.80 (95% CI 0.79–0.82; ΔAUC 0.04) | ||||||
| Validation: secondary prevention, 700 subjects (Athero-Express) | NRI: 0.17 (95% CI 0.13–0.21) | |||||
| IDI: 0.09 (95% CI 0.07–0.10) | ||||||
| Williams et al., 202234 | Derivation: secondary prevention cohorts, 813 patients (HUNT3/ARIC) | Modified aptamers (SomaScan), 5000 proteins | LASSO regression variable selection, parametric AFT final model | Composite: MI, stroke, HF hospitalization, all-cause death | 27-protein model: | |
| AUC: 0.73 (95% CI 0.72–0.74; ΔAUC 0.06) | ||||||
| Validation: primary and secondary prevention, 11 609 patients (six different cohorts) | NRI: 0.43 |
Clinical key studies using proteomics for ASCVD risk prediction.
AFT, accelerated failure time; ASCVD, atherosclerotic cardiovascular disease; AUC, area under the receiver operating curve; CABG, coronary artery bypass grafting; CHD, coronary heart disease; ELISA, enzyme-linked immunoassay; GDF-15, growth differentiation factor-15; HF, heart failure; IDI, integrated discrimination improvement; LASSO, least absolute shrinkage and selection operator; MI, myocardial infarction; NRI, net reclassification improvement; PCI, percutaneous coronary intervention; TIA, transient ischemia attack.
Ganz et al.33 were the first to develop a novel protein model for ASCVD risk prediction which was validated in an external cohort. In this study, levels of 1130 proteins were determined with a modified aptamer technique in two separate cohorts of more than 900 high-risk subjects with established ASCVD. Variables were selected using LASSO techniques, resulting in a nine-protein Cox regression model which outperformed clinical risk prediction using a refit Framingham model (net reclassification index 0.43) in predicting a composite ASCVD outcome, although overall discriminative power was limited (AUC 0.70) and included established biomarkers such as troponins. Ho et al.78 investigated the prognostic value of 85 proteins determined with a modified ELISA for a composite ASCVD outcome in 3523 subjects from the Framingham Heart Study. Only one protein (Growth differentiation factor-15; GDF-15) provided additive prognostic value when added to the Cox regression clinical risk prediction model, resulting in an AUC of 0.76. Of note, GDF-15 is highly correlated to age. Both the early studies by Ganz et al. and Ho et al. used classical Cox regression to develop their risk prediction models, which prevents accounting for non-linear relationships as well as protein–protein interactions.
More recent studies implemented more advanced machine learning techniques for the proteomics data analysis. Hoogeveen et al.31 used extreme gradient boosting to create a 50-protein model for ASCVD risk estimation in two primary prevention populations. Using proximity extension assays for 368 proteins in two parallel case–control studies of 822 and 702 patients, respectively, the protein model outperformed the clinical risk model in predicting ASCVD events. Although there was a significant benefit over the clinical model (ΔAUC 0.10), the overall discriminative value of the protein model was still limited in the validation cohort (AUC 0.70). The same proximity extension assay method for protein analysis was used by Unterhuber et al.,35 who measured 92 cardiovascular-related proteins in 1998 patients at increased ASCVD risk from the LIFE-Heart Study and validated these findings in 772 patients from the PLIC cohort. Machine learning protein prediction with extreme gradient boosting or artificial neural networks greatly outperformed both protein prediction of major adverse cardiovascular events (MACE) with regression models and traditional risk prediction with the Framingham and SCORE algorithms (AUC 0.91). Another recent study by Nurmohamed et al.32 investigated the prognostic value of 276 proteins from the proximity extension assay in two independent secondary prevention cohorts (SMART and Athero-Express). A 50-protein extreme gradient boosting machine learning model outperformed the machine learning model with clinical parameters (AUC 0.80, ΔAUC 0.04, NRI 0.17) in the validation cohort. The most recent and largest study to date, performed by Williams et al.,34 measured 5000 proteins using the modified aptamer technique. A 27-protein model was constructed after selection of parameters in the derivation cohort consisting of 813 secondary prevention patients using LASSO regression. This model was validated and compared to a refitted clinical risk model in an independent validation cohort of 11 609 patients from different studies. The final model yielded a moderate AUC of 0.73 but the protein model had a large impact on reclassification (NRI 0.43) compared to the clinical model.
Although there are marked differences in the proteins associated with (recurrent) ASCVD in the aforementioned studies, there is also a significant overlap in these top proteins. Despite different machine learning approaches, in all three studies using the proximity extension assay, NT-proBNP/brain natriuretic peptide and kidney injury molecule-1 (KIM-1) were in the top five of prognostic proteins.31,32,35 In addition, GDF-15 was in the top five in the two studies in which GDF-15 was included in the biomarker panel. Other top proteins overlapping between the studies include matrix metalloproteinase-12 (MMP-12) and adrenomedullin. The studies performed with aptamers showed somewhat more interstudy variation.33,34,78 This is possibly due to more unspecificity with the larger number of proteins analysed (>5000)—including a substantial number of proteins reflecting the same pathophysiological pathways—which will compete in ASCVD risk modelling. Matrix metalloproteinase-12 and GDF-8/11 were the only proteins that were present in both prediction models from the two studies by the group of Ganz et al. Of note, in the study by Ho et al., GDF-15 was the only protein with an additive value, which corresponds to the findings in the proximity extension assay studies. Altogether, comparison between these clinical studies is informative, but hampered by methodological and statistical differences. To unravel the importance and independent prognostic power of the proteins, further studies should be designed to address the role of specific proteins as scores involving a large number of proteins can become a barrier for clinical implementation. Instead, top proteins which were previously not studied, such as KIM-1, warrant further investigation.
Recent and key studies on lipidomics in atherosclerotic cardiovascular disease risk prediction
In parallel, a large body of evidence has linked plasma lipidomic traits to several disease phenotypes. Ceramides represent one of the best examples that have been extensively investigated to date. Increased levels of ceramides were found in acute coronary syndrom (ACS) patients,82 have been linked to insulin sensitivity in type 2 diabetes patients,83 are upregulated in chronic heart failure,84 and associated with a vulnerable plaque phenotype during intravascular ultrasound.85 In addition to ceramide species, genetic and imaging evidence has also shown that a broader set of lipid species is associated with ASCVD events and adverse coronary plaque morphology.86–89 Furthermore, different apolipoproteins have also been associated to incident ASCVD.90 Over the past years, there have been several large studies investigating the role of lipidomics in ASCVD risk prediction (Table 2).
Table 2.
Clinical key studies using lipidomics for ASCVD risk prediction
| Study and year | Study population | Lipid platform | Method of analysis | Outcome | Performance compared to clinical model | |
|---|---|---|---|---|---|---|
| Stegemann et al., 201438 | 685 subjects, community-based (Bruneck Study) | 135 lipid species, direct-infusino MS | LASSO variable selection, Cox regression | Composite: MI, ischaemic stroke, sudden cardiac death | Clinical model plus 6 lipid species: | |
| AUC: 0.75 (95% CI 0.71–0.79; ΔAUC 0.04) | ||||||
| NRI: 0.41 (95% CI 0.18–0.63) | ||||||
| Laaksonen et al., 201636 | 3384 subjects with suspected CAD or established ACS (Corogene, BECAC, and SPUM-ACS), internal validation | 4 ceramide species, LC-MS | Logistic regression | Cardiovascular death | Clinical model plus Cer(d18:1/16:0)/Cer(d18:1/24:0) ratio: | |
| AUC: 0.82 (95% CI 0.79–0.85; ΔAUC 0.09) | ||||||
| NRI: 0.17 (95% CI 0.07–0.27) | ||||||
| Alshehry et al., 201639 | Derivation: 3779 subjects with type 2 DM (case cohort from ADVANCE trial) | 310 lipid species, LC-MS | Weighted Cox regression | Cardiovascular death | Clinical model plus 4 lipid species: | |
| AUC: 0.70 (95% CI 0.68–0.71; ΔAUC 0.06) | ||||||
| Validation: 511 subjects with type 2 DM (LIPID trial) | NRI: 0.48 (95% CI 0.47–0.50) | |||||
| Havulinna et al, 201691 | 8101 subjects from general population (FINRISK 2002 cohort) | 4 ceramide species, LC-MS | Cross-validated Weibull regression | Composite: coronary heart disease, ischaemic stroke, and HF | Framingham plus ceramides: | |
| NRI: up to 0.08 | ||||||
| Wang et al., 201792 | 1017 subjects, primary prevention (case cohort from PREDIMED trial) | 4 ceramide species, LC-MS | Weighted Cox regression | Composite: MI, stroke, cardiovascular death | Clinical model plus 4-ceramide score: | |
| AUC: 0.71 (95% CI 0.66–0.73; ΔAUC 0.01) | ||||||
| NRI: 0.22 (95% CI 0.04–0.45) | ||||||
| Razquin et al., 201893 | 983 subjects, primary prevention (case cohort from PREDIMED trial) | 202 lipid species, LC-MS | Weighted Cox regression | Composite: MI, stroke, cardiovascular death | Clinical model plus lipid group: | |
| AUC: 0.71 (95% CI 0.67–0.75; ΔAUC 0.02) | ||||||
| Mundra et al., 201894 | Derivation: 5991 subjects (LIPID trial) | 342 lipid species, LC-MS | Cox regression | Cardiovascular death | Clinical model plus 7 lipid species: | |
| Validation: 3779 subjects with type 2 DM (ADVANCE trial) | AUC: 0.74 (95% CI 0.74–0.74; ΔAUC 0.02) | |||||
| NRI: 0.20 (95%CI 0.19–0.21) | ||||||
| IDI: 0.02 (95% CI 0.02–0.02) | ||||||
| Poss et al., 202095 | 674 subjects, case control (Utah Family Health Tree Program) | 32 sphingolipids, LC-MS | LASSO regression | Coronary artery disease: MI, PCI, CABG | Clinical model plus 30-sphingolipid score: | |
| AUC: 0.72 (ΔAUC 0.09) | ||||||
| NRI: 0.67 (95% CI 0.52–0.81) | ||||||
| IDI: 0.1 (95% CI 0.1–0.1) | ||||||
| Hilvo et al., 202037 | Derivation: 3882 subjects with suspected angina (WECAC) | Ceramide and phosphatidylcholine levels, LC-MS | Cox regression | Cardiovascular death | Age plus sex plus CERT2 score: | |
| AUC: 0.70 (ΔAUC 0.05; validation cohorts) | ||||||
| Validation: 5991 subjects with previous ACS (LIPID), 1206 subjects with CHD (KAROLA) | ||||||
| NRI: 0.32 (95% CI 0.14–0.50; derivation cohort) | ||||||
| Hilvo et al., 202096 | 11 222 secondary prevention subjects (STABILITY) | Ceramide and phosphatidylcholine levels, LC-MS | Cox regression | Cardiovascular death | Clinical risk model plus CERT2 score: | |
| AUC: 0.75 (95% CI 0.73–0.77; ΔAUC 0.02) | ||||||
| Ottosson et al., 202197 | 3856 primary prevention subjects (MDC-CC) | 184 lipids, direct-infusion MS | LASSO variable selection, Cox regression prediction model | Composite: coronary revascularization, MI, cardiovascular death | Clinical risk model plus eight lipid species: | |
| AUC: 0.81 (95% CI 0.78–0.84; ΔAUC 0.02) | ||||||
| Vasile et al., 202198 | 1131 subjects, community-based (REP cohort) | 4 ceramide species, LC-MS | Cox regression | Composite: stroke and MI | Ceramide score: | |
| AUC: 0.67 (95% CI 0.63–0.72; ΔAUC 0.05) | ||||||
| IDI: 0.01 (95% CI 0.00–0.02) |
ACS, acute coronary syndrome; ASCVD, atherosclerotic cardiovascular disease; AUC, area under the receiver operating curve; CABG, coronary artery bypass grafting; CHD, coronary heart disease; DM, diabetes mellitus; HF, heart failure; IDI, integrated discrimination improvement; LC, liquid chromatography; MS, mass spectrometry; LASSO, least absolute shrinkage and selection operator; MI, myocardial infarction; NRI, net reclassification improvement; PCI, percutaneous coronary intervention.
The group of Laaksonen has focused on using certain ceramide species and developing corresponding ceramide scores on top of clinical characteristics to improve future ASCVD risk prediction. Using a specifically developed and validated high-throughput LC-MS assay, four ceramide species most strongly associated with ASCVD can be reliably measured in a total analysis time of 5 min per sample.64 Thanks to the preselection resulting in this limited number of variables, these parameters could easily be added to classical regression models. In their first large-scale study, internal validation of a clinical model plus the Cer(d18:1/16:0)/Cer(d18:1/24:0) ratio resulted in an AUC of 0.82 (ΔAUC 0.09 compared to clinical model alone) and an NRI of 0.17 for prediction of cardiovascular death.36 The same group demonstrated the incremental prognostic value of the ceramide score in the FINRISK cohort.91 Following these studies, Wang et al.92 showed a slight improvement for the ceramide score over clinical risk prediction for a composite MACE endpoint in a primary prevention case cohort study comprising 1017 patients (ΔAUC 0.01, NRI 0.22). Recently, Vasile et al.98 confirmed these findings in a community-based cohort of 1131 subjects, showing additive value of the ceramide score over standard clinical risk prediction for a composite endpoint of myocardial infarction (MI) and stroke (AUC 0.67, ΔAUC 0.05). The most recent studies by the Laaksonen group described a newly developed and extended risk score containing the four ceramide species but also three phospholipids.37 This novel risk score was developed and validated in three independent cohorts comprising 11 079 patients with angina pectoris or coronary artery disease (CAD) and outperformed the standard clinical risk score in predicting cardiovascular death (AUC 0.70, ΔAUC 0.05 in validation, NRI 0.32 in derivation).37 In addition, this Cardiovascular Event Risk Test 2 (CERT2) score was used in another cohort of 11 222 stable CAD patients, where it significantly outperformed clinical characteristics in prediction of cardiovascular death (AUC 0.75, ΔAUC 0.02).96
Other studies have focused on a more lipidome-wide approach to improve risk stratification. In 2014, Stegemann et al.38 investigated 135 lipid species determined with direct-infusion MS to predict 10-year risk of CVD in a cohort of 685 subjects from the Bruneck Study. Using LASSO selection, six lipid species from five different lipid categories [TAG(54:2), PE(36:5), CE(16:1), SM(34:2), LPC(20:5), and LPC(22:6)] were found that improved ASCVD risk prediction beyond clinical characteristics using internal validation. Similarly, investigators from the Long-Term Intervention With Pravastatin in Ischemic Disease (LIPID) study showed that risk scores with respectively four and seven lipid species selected from more than 300 LS-MS determined lipid species improved prediction of cardiovascular death.39,94 In both studies, external validation showed additive clinical value with respective NRIs of 0.20 (AUC 0.74, ΔAUC 0.02) and 0.48 (AUC 0.70, ΔAUC 0.06).
Furthermore, there are several smaller single-cohort studies which deserve further attention but were not externally validated. Razquin et al.93 performed a case cohort study with 983 subjects from the Prevención con Dieta Mediterránea (PREDIMED) trial measuring 202 lipid species with LC-MS and showed that addition of several lipid species slightly improved internal prediction of MACE (AUC 0.71, ΔAUC 0.02). Poss et al.95 developed a novel 30-shingolipid score using LASSO regression which improved prediction of CAD when internally validated (0.72, ΔAUC 0.09). Finally, Ottosson et al.97 performed shotgun lipidomics (184 lipid species, direct-infusion MS) and used LASSO regression to create a risk prediction model for MACE (AUC 0.81, ΔAUC 0.02).
Implementation in atherosclerotic cardiovascular disease risk profiling
Can we implement proteomics and/or lipidomics in atherosclerotic cardiovascular disease risk algorithms?
Both proteomics and lipidomics have been extensively studied in large-scale retrospective studies, have outperformed clinical risk scores in terms of discrimination, and have shown clinically relevant reclassification improvement. In addition, for lipidomics, Hilvo et al.99 already developed a ceramide–phospholipid risk chart for use in clinical practice. Nevertheless, several limitations remain that need to be addressed before a wider clinical use of both proteomics and lipidomics in ASCVD risk algorithms can be advocated.
The most important limitation of the described biomarker studies pertains to the fact that these were all retrospective studies. Even when validated in an external cohort, the analysis remains retrospective and, perhaps even more importantly, cohort-specific; therefore, the prospective performance in different populations remains unknown. To solve this issue, controlled large studies that prospectively randomize patients according to their biomarker risk score and compare this approach to the currently used approach utilizing clinical risk scores are warranted. In such prospective studies, the algorithm cannot be calibrated; there should be a clear and interpretable risk score from a predefined (machine learning) algorithm, and there should be consensus about what risk threshold would require which treatment.
An ensuing problem to address is the choice of biomarkers to be included in such a risk score. In the described studies using proteomics or lipidomics for cardiovascular risk prediction, there are large differences in the used methodologies and assessed panels and biomarkers which limit comparison. Even between some studies using similar panels, there are major differences in the proteins associated with adverse outcome. In fact, the more biomarkers are analyzed, the larger the between-study differences in the most important biomarkers, probably due to increased competition of proteins reflecting similar pathways. Thus, for effective clinical implementation, it remains to be determined what biomarkers from which omics approach are more strongly associated with future ASCVD. In addition, ASCVD endpoints were different in the large biomarker studies, and prognostic value of biomarker profiles may depend on the type of atherosclerotic disease, e.g. myocardial infarction vs. stroke, but also on the prevention setting. The majority of studies to date has focused on coronary artery disease, but it will be important for future studies to investigate prognostic value of biomarker panels in symptomatic cerebral and peripheral artery disease.
Nevertheless, combining and comparing the large studies performed to date, there should be sufficient data to select the most important biomarkers for future prospective validation. When a panel of prognostic biomarkers can be selected, it should be feasible to combine these in one customized high-throughput assay. Such a panel can then be combined with traditional clinical risk characteristics such as age, sex, smoking status, and blood pressure, as well as apolipoprotein B or low-density lipoprotein cholesterol (LDL-C) and lipoprotein(a) [Lp(a)]—depending on the prevention setting—to provide a comprehensive ASCVD risk score.6,10,100–103 One study prospectively investigating a small set of protein and lipid biomarkers (NT-proBNP, high-sensitivity troponin, cystatin C, four ceramide species, and three phospholipid species) in a randomized controlled trial of 2000 patients is already underway (NCT04433052; www.coroprevention.eu).
Finally, implementation of large panel biomarker data in clinical risk stratification requires availability of lab equipment as well as expertise with the generated output. Currently, such omics platforms as well as the supporting industries are predominantly focused on research applications. On top of equipment and expertise availability, compliance with the strict quality regulations in diagnostic facilities handling patient care remains a hurdle to be taken.
A personalized approach for atherosclerotic cardiovascular disease prediction
The 2021 European Society of Cardiology Guidelines on cardiovascular disease prevention have underlined the importance of individualized ASCVD risk prediction.6 Although widely used, current clinical risk scores seem to fail in accurately stratifying patients at ASCVD risk. For this personalized approach, improved risk stratification beyond clinical risk factors is needed. If implemented in a high-throughput, low-cost, and reliable way, both proteomics and lipidomics have the potential to meaningfully improve ASCVD risk prediction. Combined with genetic predisposition, which could potentially be captured with polygenic risk scores,23–25 and phenotypic features of the atherosclerosis in the artery wall captured with coronary artery imaging, integration with proteomic/lipidomic data may lay the foundation for understanding the complex and dynamic nature determining an individual’s ASCVD phenotype.
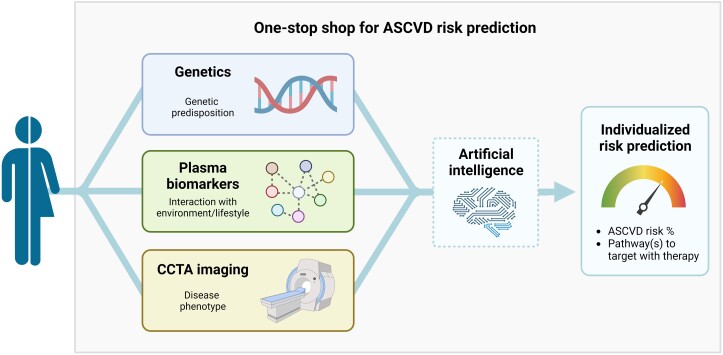
Pending validation, such a ‘one-stop shop’ model combining these three pillars for ASCVD risk prediction could be implemented in clinical practice to pursue multidimensional personalized medicine (Figure 2). In such practice, patients—during a workup for cardiovascular risk management—would undergo blood withdrawal for clinical risk factors, plasma biomarkers, and DNA analysis (polygenic risk score), followed by a CCTA scan. Using a standardized machine learning algorithm, this multidimensional datum can be translated into an individualized ASCVD risk prediction. Most importantly, such individualized, multidimensional ASCVD risk prediction should be reported in a simple risk score to enable clinical implementation. Furthermore, it is tempting to speculate that incorporation of pathway analysis in the omics sets—when feasible—may facilitate identification of most affected pathways underlying an individual’s ASCVD risk, which could assist in defining the best therapeutic strategies (thus adding anti-inflammatory, anti-thrombotic, and/or lipid-lowering agents).28 In relatively young primary prevention patients (e.g. below 55 years), the combination of genetic testing, measurement of plasma biomarkers, and CCTA imaging is likely to identify high-risk patients which would not have qualified for treatment following the SCORE2 charts. In contrast, in patients with a low polygenic/biomarker risk score and absence of coronary atherosclerosis, further treatment could be withheld. In patients with a high polygenic/biomarker risk but absence of coronary atherosclerosis on imaging, strategy will depend on the age of the patient. Younger patients with a high polygenic/biomarker risk are likely to be treated, given their lifetime ASCVD risk increase. Conversely, older patients with absence of coronary disease on imaging (e.g. above 65 years) may not require treatment in view of their low ASCVD risk considering the ‘warranty period’ of a normal cardiac CT scan.104 Given the profound cost reductions in these diagnostic approaches over the last years combined with the high costs of novel therapeutics, this individualized risk-guided strategy is likely to be cost-effective. It should, however, be taken into account that the endeavour to perform a prospective clinical utility study of such an integrated approach will require an enormous investment.
Figure 2.
A one-stop shop for future ASCVD risk prediction. A personalized atherosclerotic cardiovascular disease risk prediction in a one-stop shop can—in addition to clinical risk factors—incorporate a patient’s genetic predisposition, capture environmental and lifestyle factors in interaction with genetics using plasma biomarkers, and can define the actual phenotype of disease using coronary computed tomography angiography imaging. In addition, the most ‘relevant’ pathways contributing to the cardiovascular risk in specific individuals could be identified and subsequently treated with specific therapies, depending on the specific risk factor signature (e.g. anti-thrombotic, anti-inflammatory, and other). Created with BioRender.com. ASCVD, atherosclerotic cardiovascular disease; CCTA, coronary computed tomography angiography.
For secondary prevention, the same approach could be used after a cardiovascular event to identify patients with a higher residual risk despite guideline-based risk-lowering therapies. In patients with an unidentified high polygenic, biomarker, and/or imaging-derived risk, it can be considered to apply lower target levels for established risk factors (e.g. LDL-C), as is currently advocated for e.g. primary prevention patients with elevated Lp(a), or to prescribe additional preventive therapies, depending on the specific risk factor signature (e.g. anti-thrombotic, anti-inflammatory, and other).6
Pending the analytical and machine learning hurdles to take for such a sophisticated one-stop shop risk score, interim solutions such as proposed by Hilvo et al.105—a risk chart combining imaging, lipidomic, and clinical data—may be implemented in the short-term.
Conclusions
Plasma proteomics and lipidomics hold a major promise to improve the limited prediction of individual ASCVD risk using currently available algorithms (Graphical Abstract). Using several techniques, both plasma proteins and lipids can be reliably assessed using high-throughput assays and have established additive value for ASCVD risk stratification, albeit retrospectively. There is a lack of prospective proteomics and lipidomics studies, and selection of prognostic biomarkers remains problematic due to substantial interstudy differences. Pending analytical and clinical validation, clinical utility studies investigating the value of biomarker panels when applied in clinical practice are eagerly awaited. Ultimately, prognostic plasma biomarkers could be combined with genetic risk scores and coronary artery imaging to capture the complex, multidimensional atherosclerosis process underlying an individual’s ASCVD risk.
Contributor Information
Nick S Nurmohamed, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands; Department of Cardiology, Amsterdam University Medical Centers, Vrije Universiteit Amsterdam, De Boelelaan 1117, 1081 HV Amsterdam, The Netherlands.
Jordan M Kraaijenhof, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands.
Manuel Mayr, School of Cardiovascular and Metabolic Medicine & Science, King’s College London, Strand, London WC2R 2LS, UK; Department of Internal Medicine II, Division of Cardiology, Medical University of Vienna, Währinger Gürtel, 18-201090 Vienna, Austria.
Stephen J Nicholls, Victorian Heart Institute, Monash University, 631 Blackburn Rd, Clayton, VIC 3168, Australia.
Wolfgang Koenig, Deutsches Herzzentrum München, Technische Universität München, Lazarettstraße 36, 80636 München, Germany; German Centre for Cardiovascular Research (DZHK e.V.), partner site Munich Heart Alliance, Pettenkoferstr. 8a & 9, 80336 Munich, Germany; Institute of Epidemiology and Medical Biometry, University of Ulm, Helmholtzstr. 22, 89081 Ulm, Germany.
Alberico L Catapano, Department of Pharmacological and Biomolecular Sciences, University of Milan, Via Balzaretti 9, 20133 Milan, Italy; IRCCS Multimedica, Via Milanese, 300, 20099 Sesto San Giovanni (MI), Italy.
Erik S G Stroes, Department of Vascular Medicine, Amsterdam University Medical Centers, University of Amsterdam, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands.
Data Availability
No new data were generated or analysed in support of this research.
Funding
This work was supported by a European Research Area Network on Cardiovascular Diseases (ERA-CVD) grant (ERA-CVD JTC2017). M.M. is a British Heart Foundation (BHF) Chair Holder (CH/16/3/32406) with BHF programme grant support (RG/F/21/110053). M.M. is also supported by the Leducq Foundation (18CVD02) and the VASCage—Research Center on Vascular Ageing and Stroke (No. 868624).
References
- 1. Yusuf S, Rangarajan S, Teo K, Islam S, Li W, Liu L, et al. . Cardiovascular risk and events in 17 low-, middle-, and high-income countries. N Engl J Med 2014;371:818–827. 10.1056/NEJMoa1311890 [DOI] [PubMed] [Google Scholar]
- 2. Fernández-Friera L, Fuster V, López-Melgar B, Oliva B, García-Ruiz JM, Mendiguren J, et al. . Normal LDL-cholesterol levels are associated with subclinical atherosclerosis in the absence of risk factors. J Am Coll Cardiol 2017;70:2979–2991. 10.1016/j.jacc.2017.10.024 [DOI] [PubMed] [Google Scholar]
- 3. Jernberg T, Hasvold P, Henriksson M, Hjelm H, Thuresson M, Janzon M. Cardiovascular risk in post-myocardial infarction patients: nationwide real world data demonstrate the importance of a long-term perspective. Eur Heart J 2015;36:1163–1170. 10.1093/eurheartj/ehu505 [DOI] [PubMed] [Google Scholar]
- 4. Mortensen MB, Nordestgaard BG, Afzal S, Falk E. ACC/AHA guidelines superior to ESC/EAS guidelines for primary prevention with statins in non-diabetic Europeans: the Copenhagen General Population Study. Eur Heart J 2017;38:586–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kaasenbrood L, Boekholdt SM, Van Der GY, Ray KK, Peters RJG, Kastelein JJP, et al. . Distribution of estimated 10-year risk of recurrent vascular events and residual risk in a secondary prevention population. Circulation 2016;134:1419–1429. 10.1161/CIRCULATIONAHA.116.021314 [DOI] [PubMed] [Google Scholar]
- 6. Visseren FLJ, MacH F, Smulders YM, Carballo D, Koskinas KC, Bäck M, et al. . 2021 ESC guidelines on cardiovascular disease prevention in clinical practice. Eur Heart J 2021;42:3227–3337. 10.1093/eurheartj/ehab484 [DOI] [PubMed] [Google Scholar]
- 7. Goff DC, Lloyd-Jones DM, Bennett G, Coady S, D’Agostino RB, Gibbons R, et al. . 2013 ACC/AHA guideline on the assessment of cardiovascular risk: a report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines. Circulation 2014;129:S49–S73. 10.1161/01.cir.0000437741.48606.98 [DOI] [PubMed] [Google Scholar]
- 8. D’Agostino RB, Vasan RS, Pencina MJ, Wolf PA, Cobain M, Massaro JM, et al. . General cardiovascular risk profile for use in primary care: the Framingham Heart Study. Circulation 2008;117:743–753. 10.1161/CIRCULATIONAHA.107.699579 [DOI] [PubMed] [Google Scholar]
- 9. JBS3 Board . Joint British Societies’ consensus recommendations for the prevention of cardiovascular disease (JBS3). Heart 2014;100:ii1–ii67. 10.1136/heartjnl-2014-305693 [DOI] [PubMed] [Google Scholar]
- 10. Hageman SHJ, McKay AJ, Ueda P, Gunn LH, Jernberg T, Hagström E, et al. . Estimation of recurrent atherosclerotic cardiovascular event risk in patients with established cardiovascular disease: the updated SMART2 algorithm. Eur Heart J 2022;43:1715–1727. 10.1093/eurheartj/ehac056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ray KK, Wright RS, Kallend D, Koenig W, Leiter LA, Raal FJ, et al. . Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med 2020;382:1507–1519. 10.1056/NEJMoa1912387 [DOI] [PubMed] [Google Scholar]
- 12. Raal FJ, Kallend D, Ray KK, Turner T, Koenig W, Wright RS, et al. . Inclisiran for the treatment of heterozygous familial hypercholesterolemia. N Engl J Med 2020;382:1520–1530. 10.1056/NEJMoa1913805 [DOI] [PubMed] [Google Scholar]
- 13. Packer M, Anker SD, Butler J, Filippatos G, Pocock SJ, Carson P, et al. . Cardiovascular and renal outcomes with empagliflozin in heart failure. N Engl J Med 2020;383:1413–1424. 10.1056/NEJMoa2022190 [DOI] [PubMed] [Google Scholar]
- 14. Wiviott SD, Raz I, Bonaca MP, Mosenzon O, Kato ET, Cahn A, et al. . Dapagliflozin and cardiovascular outcomes in type 2 diabetes. N Engl J Med 2019;380:347–357. 10.1056/NEJMoa1812389 [DOI] [PubMed] [Google Scholar]
- 15. Neal B, Perkovic V, Mahaffey KW, de Zeeuw D, Fulcher G, Erondu N, et al. . Canagliflozin and cardiovascular and renal events in type 2 diabetes. N Engl J Med 2017;377:644–657. 10.1056/NEJMoa1611925 [DOI] [PubMed] [Google Scholar]
- 16. Husain M, Birkenfeld AL, Donsmark M, Dungan K, Eliaschewitz FG, Franco DR, et al. . Oral semaglutide and cardiovascular outcomes in patients with type 2 diabetes. N Engl J Med 2019;381:841–851. 10.1056/NEJMoa1901118 [DOI] [PubMed] [Google Scholar]
- 17. Marso SP, Daniels GH, Frandsen KB, Kristensen P, Mann JFE, Nauck MA, et al. . Liraglutide and cardiovascular outcomes in type 2 diabetes. Drug Ther Bull 2016;54:101. [Google Scholar]
- 18. Ray KK, Bays HE, Catapano AL, Lalwani ND, Bloedon LT, Sterling LR, et al. . Safety and efficacy of bempedoic acid to reduce LDL cholesterol. N Engl J Med 2019;380:1022–1032. 10.1056/NEJMoa1803917 [DOI] [PubMed] [Google Scholar]
- 19. Goldberg AC, Leiter LA, Stroes ESG, Baum SJ, Hanselman JC, Bloedon LAT, et al. . Effect of bempedoic acid vs placebo added to maximally tolerated statins on low-density lipoprotein cholesterol in patients at high risk for cardiovascular disease: the CLEAR wisdom randomized clinical trial. JAMA 2019;322:1780–1788. 10.1001/jama.2019.16585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nidorf SM, Fiolet ATL, Mosterd A, Eikelboom JW, Schut A, Opstal TSJ, et al. . Colchicine in patients with chronic coronary disease. N Engl J Med 2020;383:1838–1847. 10.1056/NEJMoa2021372 [DOI] [PubMed] [Google Scholar]
- 21. Ridker PM, Everett BM, Thuren T, MacFadyen JG, Chang WH, Ballantyne C, et al. . Antiinflammatory therapy with canakinumab for atherosclerotic disease. N Engl J Med 2017;377:1119–1131. 10.1056/NEJMoa1707914 [DOI] [PubMed] [Google Scholar]
- 22. Varghese MS, Liu CL, Kazi DS. The price of progress: cost, access, and adoption of novel cardiovascular drugs in clinical practice. Curr Cardiol Rep 2021;23:163. 10.1007/s11886-021-01598-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Khera A V, Chaffin M, Aragam KG, Haas ME, Roselli C, Choi SH, et al. . Genome-wide polygenic scores for common diseases identify individuals with risk equivalent to monogenic mutations. Nat Genet 2018;50:1219–1224. 10.1038/s41588-018-0183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Neumann JT, Riaz M, Bakshi A, Polekhina G, Thao LTP, Nelson MR, et al. . Prognostic value of a polygenic risk score for coronary heart disease in individuals aged 70 years and older. Circ Genomic Precis Med 2022;15:E003429. 10.1161/CIRCGEN.121.003429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Maamari DJ, Brockman DG, Aragam K, Pelletier RC, Folkerts E, Neben CL, et al. . Clinical implementation of combined monogenic and polygenic risk disclosure for coronary artery disease. JACC Adv 2022;1:100068. 10.1016/j.jacadv.2022.100068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Abdelrahman KM, Chen MY, Dey AK, Virmani R, Finn A V, Khamis RY, et al. . Coronary computed tomography angiography from clinical uses to emerging technologies: JACC state-of-the-art review. J Am Coll Cardiol 2020;76:1226–1243. 10.1016/j.jacc.2020.06.076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Griffin WF, Choi AD, Riess JS, Marques H, Chang H-J, Choi JH, et al. . AI evaluation of stenosis on coronary CT angiography, comparison with quantitative coronary angiography and fractional flow reserve. JACC Cardiovasc Imaging 2023;16:193–205. 10.1016/j.jcmg.2021.10.020. [Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 28. Ridker PM. Clinician’s guide to reducing inflammation to reduce atherothrombotic risk: JACC review topic of the week. J Am Coll Cardiol 2018;72:3320–3331. 10.1016/j.jacc.2018.06.082 [DOI] [PubMed] [Google Scholar]
- 29. Nambi V, Ballantyne CM, Hoogeveen RC, Agarwal SK, Panagiotakos DB, Wannamethee SG, et al. . Natriuretic peptides and integrated risk assessment for cardiovascular disease: an individual-participant-data meta-analysis. Lancet Diabetes Endocrinol 2016;4:840–849. 10.1016/S2213-8587(16)30196-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Farmakis D, Mueller C, Apple FS. High-sensitivity cardiac troponin assays for cardiovascular risk stratification in the general population. Eur Heart J 2020;41:4050–4056. 10.1093/eurheartj/ehaa083 [DOI] [PubMed] [Google Scholar]
- 31. Hoogeveen RM, Pereira JPB, Nurmohamed NS, Zampoleri V, Bom MJ, Baragetti A, et al. . Improved cardiovascular risk prediction using targeted plasma proteomics in primary prevention. Eur Heart J 2020;41:3998–4007. 10.1093/eurheartj/ehaa648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nurmohamed NS, Belo Pereira JP, Hoogeveen RM, Kroon J, Kraaijenhof JM, Waissi F, et al. . Targeted proteomics improves cardiovascular risk prediction in secondary prevention. Eur Heart J 2022;43:1569–1577. 10.1093/eurheartj/ehac055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ganz P, Heidecker B, Hveem K, Jonasson C, Kato S, Segal MR, et al. . Development and validation of a protein-based risk score for cardiovascular outcomes among patients with stable coronary heart disease. JAMA 2016;315:2532–2541. 10.1001/jama.2016.5951 [DOI] [PubMed] [Google Scholar]
- 34. Williams SA, Ostroff R, Hinterberg MA, Coresh J, Ballantyne CM, Matsushita K, et al. . A proteomic surrogate for cardiovascular outcomes that is sensitive to multiple mechanisms of change in risk. Sci Transl Med 2022;14:eabj9625. 10.1126/scitranslmed.abj9625 [DOI] [PubMed] [Google Scholar]
- 35. Unterhuber M, Kresoja KP, Rommel KP, Besler C, Baragetti A, Klöting N, et al. . Proteomics-enabled deep learning machine algorithms can enhance prediction of mortality. J Am Coll Cardiol 2021;78:1621–1631. 10.1016/j.jacc.2021.08.018 [DOI] [PubMed] [Google Scholar]
- 36. Laaksonen R, Ekroos K, Sysi-Aho M, Hilvo M, Vihervaara T, Kauhanen D, et al. . Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J 2016;37:1967–1976. 10.1093/eurheartj/ehw148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hilvo M, Meikle PJ, Pedersen ER, Tell GS, Dhar I, Brenner H, et al. . Development and validation of a ceramide- and phospholipid-based cardiovascular risk estimation score for coronary artery disease patients. Eur Heart J 2020;41:371–380. 10.1093/eurheartj/ehz387 [DOI] [PubMed] [Google Scholar]
- 38. Stegemann C, Pechlaner R, Willeit P, Langley SR, Mangino M, Mayr U, et al. . Lipidomics profiling and risk of cardiovascular disease in the prospective population-based Bruneck study. Circulation 2014;129:1821–1831. 10.1161/CIRCULATIONAHA.113.002500 [DOI] [PubMed] [Google Scholar]
- 39. Alshehry ZH, Mundra PA, Barlow CK, Mellett NA, Wong G, McConville MJ, et al. . Plasma lipidomic profiles improve on traditional risk factors for the prediction of cardiovascular events in type 2 diabetes mellitus. Circulation 2016;134:1637–1650. 10.1161/CIRCULATIONAHA.116.023233 [DOI] [PubMed] [Google Scholar]
- 40. Swinbanks D. Australia backs innovation, shuns telescope. Nature 1995;378:653–653. 10.1038/378653a0 [DOI] [PubMed] [Google Scholar]
- 41. Schwenk JM, Omenn GS, Sun Z, Campbell DS, Baker MS, Overall CM, et al. . The human plasma proteome draft of 2017: building on the human plasma PeptideAtlas from mass spectrometry and complementary assays. J Proteome Res 2017;16:4299–4310. 10.1021/acs.jproteome.7b00467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, et al. . Lipidomics reveals a remarkable diversity of lipids in human plasma1. J Lipid Res 2010;51:3299–3305. 10.1194/jlr.M009449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Tabassum R, Ripatti S. Integrating lipidomics and genomics: emerging tools to understand cardiovascular diseases. Cell Mol Life Sci 2021;78:2565–2584. 10.1007/s00018-020-03715-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lindsey ML, Mayr M, Gomes A V, Delles C, Arrell DK, Murphy AM, et al. . Transformative impact of proteomics on cardiovascular health and disease: a scientific statement from the American Heart Association. Circulation 2015;132:852–872. 10.1161/CIR.0000000000000226 [DOI] [PubMed] [Google Scholar]
- 45. Blume JE, Manning WC, Troiano G, Hornburg D, Figa M, Hesterberg L, et al. . Rapid, deep and precise profiling of the plasma proteome with multi-nanoparticle protein corona. Nat Commun 2020;11:3662. 10.1038/s41467-020-17033-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Tognetti M, Sklodowski K, Müller S, Kamber D, Muntel J, Bruderer R, et al. . Biomarker candidates for tumors identified from deep-profiled plasma stem predominantly from the low abundant area. J Proteome Res 2022;21:1718–1735. 10.1021/acs.jproteome.2c00122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Griss J, Perez-Riverol Y, Lewis S, Tabb DL, Dianes JA, Del-Toro N, et al. . Recognizing millions of consistently unidentified spectra across hundreds of shotgun proteomics datasets. Nat Methods 2016;13:651–656. 10.1038/nmeth.3902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Assarsson E, Lundberg M, Holmquist G, Björkesten J, Thorsen SB, Ekman D, et al. . Homogenous 96-plex PEA immunoassay exhibiting high sensitivity, specificity, and excellent scalability. PLoS One 2014;9:e95192. 10.1371/journal.pone.0095192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Haslam DE, Li J, Dillon ST, Gu X, Cao Y, Zeleznik OA, et al. . Stability and reproducibility of proteomic profiles in epidemiological studies: comparing the Olink and SOMAscan platforms. Proteomics 2022;22:2100170. 10.1002/pmic.202100170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Brody E, Gold L, Mehan M, Ostroff R, Rohloff J, Walker J, et al. . Life’s simple measures: unlocking the proteome. J Mol Biol 2012;422:595–606. 10.1016/j.jmb.2012.06.021 [DOI] [PubMed] [Google Scholar]
- 51. Kim CH, Tworoger SS, Stampfer MJ, Dillon ST, Gu X, Sawyer SJ, et al. . Stability and reproducibility of proteomic profiles measured with an aptamer-based platform. Sci Rep 2018;8:8382. 10.1038/s41598-018-26640-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Pietzner M, Wheeler E, Carrasco-Zanini J, Kerrison ND, Oerton E, Koprulu M, et al. . Synergistic insights into human health from aptamer- and antibody-based proteomic profiling. Nat Commun 2021;12:6822. 10.1038/s41467-021-27164-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Katz DH, Robbins JM, Deng S, Tahir UA, Bick AG, Pampana A, et al. . Proteomic profiling platforms head to head: leveraging genetics and clinical traits to compare aptamer- and antibody-based methods. Sci Adv 2022;8:eabm5164. 10.1126/sciadv.abm5164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Corthals CL, Wasinger VC, Hochstrasser DF, Sanchez JC. The dynamic range of protein expression: a challenge for proteomic research. Electrophoresis 2000;21:1104–1115. [DOI] [PubMed] [Google Scholar]
- 55. Ochsner UA, Green LS, Rice TP, Olivas E, Janjic N, Katilius E. Targeting unique epitopes on highly similar proteins GDF-11 and GDF-8 with modified DNA aptamers. Biochemistry 2019;58:4632–4640. 10.1021/acs.biochem.9b00760 [DOI] [PubMed] [Google Scholar]
- 56. Raffield LM, Dang H, Pratte KA, Jacobson S, Gillenwater LA, Ampleford E, et al. . Comparison of proteomic assessment methods in multiple cohort studies. Proteomics 2020;20:1900278. 10.1002/pmic.201900278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Lasseter HC, Provost AC, Chaby LE, Daskalakis NP, Haas M, Jeromin A. Cross-platform comparison of highly sensitive immunoassay technologies for cytokine markers: platform performance in post-traumatic stress disorder and Parkinson’s disease. Cytokine X 2020;2:100027. 10.1016/j.cytox.2020.100027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Juncker D, Bergeron S, Laforte V, Li H. Cross-reactivity in antibody microarrays and multiplexed sandwich assays: shedding light on the dark side of multiplexing. Curr Opin Chem Biol 2014;18:29–37. 10.1016/j.cbpa.2013.11.012 [DOI] [PubMed] [Google Scholar]
- 59. Aksel T, Qian H, Hao P, Indermuhle PF, Inman C, Paul S, et al. . High-density and scalable protein arrays for single-molecule proteomic studies. bioRxiv 2022. 2022.05.02.490328. [Google Scholar]
- 60. Doerr A. DIA mass spectrometry. Nat Methods 2015;12:35. 10.1038/nmeth.3234 [DOI] [Google Scholar]
- 61. Joshi A, Rienks M, Theofilatos K, Mayr M. Systems biology in cardiovascular disease: a multiomics approach. Nat Rev Cardiol 2021;18:313–330. 10.1038/s41569-020-00477-1 [DOI] [PubMed] [Google Scholar]
- 62. Whitehouse CM, Dreyer RN, Yamashita M, Fenn JB. Electrospray interface for liquid chromatographs and mass spectrometers. Anal Chem 1985;57:675–679. 10.1021/ac00280a023 [DOI] [PubMed] [Google Scholar]
- 63. Hsu FF. Mass spectrometry-based shotgun lipidomics—a critical review from the technical point of view. Anal Bioanal Chem 2018;410:6387–6409. 10.1007/s00216-018-1252-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kauhanen D, Sysi-Aho M, Koistinen KM, Laaksonen R, Sinisalo J, Ekroos K. Development and validation of a high-throughput LC–MS/MS assay for routine measurement of molecular ceramides. Anal Bioanal Chem 2016;408:3475–3483. 10.1007/s00216-016-9425-z [DOI] [PubMed] [Google Scholar]
- 65. Peduzzi P, Concato J, Kemper E, Holford TR, Feinstem AR. A simulation study of the number of events per variable in logistic regression analysis. J Clin Epidemiol 1996;49:1373–1379. 10.1016/S0895-4356(96)00236-3 [DOI] [PubMed] [Google Scholar]
- 66. Tibshirani R. The LASSO method for variable selection in the Cox model. Stat Med 1997;16:385–395. [DOI] [PubMed] [Google Scholar]
- 67. Tibshirani R. Regression shrinkage and selection via the lasso: a retrospective. J R Stat Soc Ser B Stat Methodol 2011;73:273–282. 10.1111/j.1467-9868.2011.00771.x [DOI] [Google Scholar]
- 68. Al’Aref SJ, Anchouche K, Singh G, Slomka PJ, Kolli KK, Kumar A, et al. . Clinical applications of machine learning in cardiovascular disease and its relevance to cardiac imaging. Eur Heart J 2019;40:1975–1986. 10.1093/eurheartj/ehy404 [DOI] [PubMed] [Google Scholar]
- 69. Meinshausen N, Bühlmann P. Stability selection. J R Stat Soc Ser B Stat Methodol 2010;72:417–473. 10.1111/j.1467-9868.2010.00740.x [DOI] [Google Scholar]
- 70. Cattelani L, Fortino V. Improved NSGA-II algorithms for multi-objective biomarker discovery. Bioinformatics 2022;38:ii20–ii26. 10.1093/bioinformatics/btac463 [DOI] [PubMed] [Google Scholar]
- 71. McCulloch WS, Pitts W. A logical calculus of the ideas immanent in nervous activity. Bull Math Biophys 1943;5:115–133. 10.1007/BF02478259 [DOI] [PubMed] [Google Scholar]
- 72. Chen T, Guestrin C. XGBoost: a scalable tree boosting system. Proceedings of the ACM SIGKDD International Conference on Knowledge Discovery and Data Mining.https://dl.acm.org/doi/pdf/10.1145/2939672.2939785
- 73. Quer G, Arnaout R, Henne M, Arnaout R. Machine learning and the future of cardiovascular care: JACC state-of-the-art review. J Am Coll Cardiol 2021;77:300–313. 10.1016/j.jacc.2020.11.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, et al. . Assessing the performance of prediction models: a framework for traditional and novel measures. Epidemiology 2010;21:128–138. 10.1097/EDE.0b013e3181c30fb2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pencina MJ, D’Agostino RB, Steyerberg EW. Extensions of net reclassification improvement calculations to measure usefulness of new biomarkers. Stat Med 2011;30:11–21. 10.1002/sim.4085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Pencina MJ, Steyerberg EW, D’Agostino RB. Net reclassification index at event rate: properties and relationships. Stat Med 2017;36:4455–4467. 10.1002/sim.7041 [DOI] [PubMed] [Google Scholar]
- 77. Yin X, Subramanian S, Hwang SJ, O’Donnell CJ, Fox CS, Courchesne P, et al. . Protein biomarkers of new-onset cardiovascular disease: prospective study from the systems approach to biomarker research in cardiovascular disease initiative. Arterioscler Thromb Vasc Biol 2014;34:939–945. 10.1161/ATVBAHA.113.302918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Ho JE, Lyass A, Courchesne P, Chen G, Liu C, Yin X, et al. . Protein biomarkers of cardiovascular disease and mortality in the community. J Am Heart Assoc 2018;7:e008108. 10.1161/JAHA.117.008108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Bom MJ, Levin E, Driessen RS, Danad I, Van Kuijk CC, van Rossum AC, et al. . Predictive value of targeted proteomics for coronary plaque morphology in patients with suspected coronary artery disease. EBioMedicine 2019;39:109–117. 10.1016/j.ebiom.2018.12.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Lind L, Siegbahn A, Lindahl B, Stenemo M, Sundström J, Ärnlöv J. Discovery of new risk markers for ischemic stroke using a novel targeted proteomics chip. Stroke 2015;46:3340–3347. 10.1161/STROKEAHA.115.010829 [DOI] [PubMed] [Google Scholar]
- 81. Nowak C, Carlsson AC, Östgren CJ, Nyström FH, Alam M, Feldreich T, et al. . Multiplex proteomics for prediction of major cardiovascular events in type 2 diabetes. Diabetologia 2018;61:1748–1757. 10.1007/s00125-018-4641-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Pan W, Yu J, Shi R, Yan L, Yang T, Li Y, et al. . Elevation of ceramide and activation of secretory acid sphingomyelinase in patients with acute coronary syndromes. Coron Artery Dis 2014;25:230–235. 10.1097/MCA.0000000000000079 [DOI] [PubMed] [Google Scholar]
- 83. Boon J, Hoy AJ, Stark R, Brown RD, Meex RC, Henstridge DC, et al. . Ceramides contained in LDL are elevated in type 2 diabetes and promote inflammation and skeletal muscle insulin resistance. Diabetes 2013;62:401–410. 10.2337/db12-0686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Yu J, Pan W, Shi R, Yang T, Li Y, Yu G, et al. . Ceramide is upregulated and associated with mortality in patients with chronic heart failure. Can J Cardiol 2015;31:357–363. 10.1016/j.cjca.2014.12.007 [DOI] [PubMed] [Google Scholar]
- 85. Cheng JM, Suoniemi M, Kardys I, Vihervaara T, de Boer SPM, Akkerhuis KM, et al. . Plasma concentrations of molecular lipid species in relation to coronary plaque characteristics and cardiovascular outcome: results of the ATHEROREMO-IVUS study. Atherosclerosis 2015;243:560–566. 10.1016/j.atherosclerosis.2015.10.022 [DOI] [PubMed] [Google Scholar]
- 86. Tabassum R, Rämö JT, Ripatti P, Koskela JT, Kurki M, Karjalainen J, et al. . Genetic architecture of human plasma lipidome and its link to cardiovascular disease. Nat Commun 2019;10:4329. 10.1038/s41467-019-11954-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Bellis C, Kulkarni H, Mamtani M, Kent JW, Wong G, Weir JM, et al. . Human plasma lipidome is pleiotropically associated with cardiovascular risk factors and death. Circ Cardiovasc Genet 2014;7:854–863. 10.1161/CIRCGENETICS.114.000600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Ellims AH, Wong G, Weir JM, Lew P, Meikle PJ, Taylor AJ. Plasma lipidomic analysis predicts non-calcified coronary artery plaque in asymptomatic patients at intermediate risk of coronary artery disease. Eur Heart J Cardiovasc Imaging 2014;15:908–916. 10.1093/ehjci/jeu033 [DOI] [PubMed] [Google Scholar]
- 89. Bodini A, Michelucci E, Di Giorgi N, Caselli C, Signore G, Neglia D, et al. . Predictive added value of selected plasma lipids to a re-estimated minimal risk tool. Front Cardiovasc Med 2021;8:682785. 10.3389/fcvm.2021.682785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Pechlaner R, Tsimikas S, Yin X, Willeit P, Baig F, Santer P, et al. . Very-low-density lipoprotein–associated apolipoproteins predict cardiovascular events and are lowered by inhibition of APOC-III. J Am Coll Cardiol 2017;69:789–800. 10.1016/j.jacc.2016.11.065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Havulinna AS, Sysi-Aho M, Hilvo M, Kauhanen D, Hurme R, Ekroos K, et al. . Circulating ceramides predict cardiovascular outcomes in the population-based FINRISK 2002 cohort. Arterioscler Thromb Vasc Biol 2016;36:2424–2430. 10.1161/ATVBAHA.116.307497 [DOI] [PubMed] [Google Scholar]
- 92. Wang DD, Toledo E, Hruby A, Rosner BA, Willett WC, Sun Q, et al. . Plasma ceramides, Mediterranean diet, and incident cardiovascular disease in the PREDIMED trial (Prevención con Dieta Mediterránea). Circulation 2017;135:2028–2040. 10.1161/CIRCULATIONAHA.116.024261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Razquin C, Liang L, Toledo E, Clish CB, Ruiz-Canela M, Zheng Y, et al. . Plasma lipidome patterns associated with cardiovascular risk in the PREDIMED trial: a case-cohort study. Int J Cardiol 2018;253:126–132. 10.1016/j.ijcard.2017.10.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Mundra PA, Barlow CK, Nestel PJ, Barnes EH, Kirby A, Thompson P, et al. . Large-scale plasma lipidomic profiling identifies lipids that predict cardiovascular events in secondary prevention. JCI insight 2018;3:e121326. 10.1172/jci.insight.121326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Poss AM, Maschek JA, Cox JE, Hauner BJ, Hopkins PN, Hunt SC, et al. . Machine learning reveals serum sphingolipids as cholesterol-independent biomarkers of coronary artery disease. J Clin Invest 2020;130:1363–1376. 10.1172/JCI131838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Hilvo M, Wallentin L, Lakic TG, Held C, Kauhanen D, Jylhä A, et al. . Prediction of residual risk by ceramide-phospholipid score in patients with stable coronary heart disease on optimal medical therapy. J Am Heart Assoc 2020;9:e015258. 10.1161/JAHA.119.015258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Ottosson F, Emami Khoonsari P, Gerl MJ, Simons K, Melander O, Fernandez C. A plasma lipid signature predicts incident coronary artery disease. Int J Cardiol 2021;331:249–254. 10.1016/j.ijcard.2021.01.059 [DOI] [PubMed] [Google Scholar]
- 98. Vasile VC, Meeusen JW, Medina Inojosa JR, Donato LJ, Scott CG, Hyun MS, et al. . Ceramide scores predict cardiovascular risk in the community. Arterioscler Thromb Vasc Biol 2021;41:1558–1569. 10.1161/ATVBAHA.120.315530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Hilvo M, Jylhä A, Lääperi M, Jousilahti P, Laaksonen R. Absolute and relative risk prediction in cardiovascular primary prevention with a modified SCORE chart incorporating ceramide-phospholipid risk score and diabetes mellitus. Eur Heart J Open 2021;1:oeab010. 10.1093/ehjopen/oeab010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ference BA, Ginsberg HN, Graham I, Ray KK, Packard CJ, Bruckert E, et al. . Low-density lipoproteins cause atherosclerotic cardiovascular disease. 1. Evidence from genetic, epidemiologic, and clinical studies. A consensus statement fromthe European Atherosclerosis Society Consensus Panel. Eur Heart J 2017;38:2459–2472. 10.1093/eurheartj/ehx144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Mach F, Baigent C, Catapano AL, Koskinas KC, Casula M, Badimon L, et al. . 2019 ESC/EAS guidelines for the management of dyslipidaemias: lipid modification to reduce cardiovascular risk. Eur Heart J 2020;41:111–188. 10.1093/eurheartj/ehz455 [DOI] [PubMed] [Google Scholar]
- 102. Nurmohamed NS, Kaiser Y, Schuitema PCE, Ibrahim S, Nierman M, Fischer JC, et al. . Finding very high lipoprotein(a): the need for routine assessment. Eur J Prev Cardiol 2022;29:769–776. 10.1093/eurjpc/zwab167 [DOI] [PubMed] [Google Scholar]
- 103. Kronenberg F, Mora S, Stroes ESG, Ference BA, Arsenault BJ, Berglund L, et al. . Lipoprotein(a) in atherosclerotic cardiovascular disease and aortic stenosis: a European Atherosclerosis Society consensus statement. Eur Heart J 2022;43:3925–3946. 10.1093/eurheartj/ehac361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Dzaye O, Dardari ZA, Cainzos-Achirica M, Blankstein R, Agatston AS, Duebgen M, et al. . Warranty period of a calcium score of zero: comprehensive analysis from MESA. JACC Cardiovasc Imaging 2021;14:990–1002. 10.1016/j.jcmg.2020.06.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Hilvo M, Lääperi M, Jylhä A, Kleber ME, Hurme R, Scharnagl H, et al. . Prior myocardial infarction, coronary artery disease extent, diabetes mellitus, and CERT2 score for risk stratification in stable coronary artery disease. Eur J Prev Cardiol 2022;29:E159–E162. 10.1093/eurjpc/zwab122 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
No new data were generated or analysed in support of this research.