Figure 8. Modulation of SNARE complex assembly by SMN and the G470R variant of Hspa8.
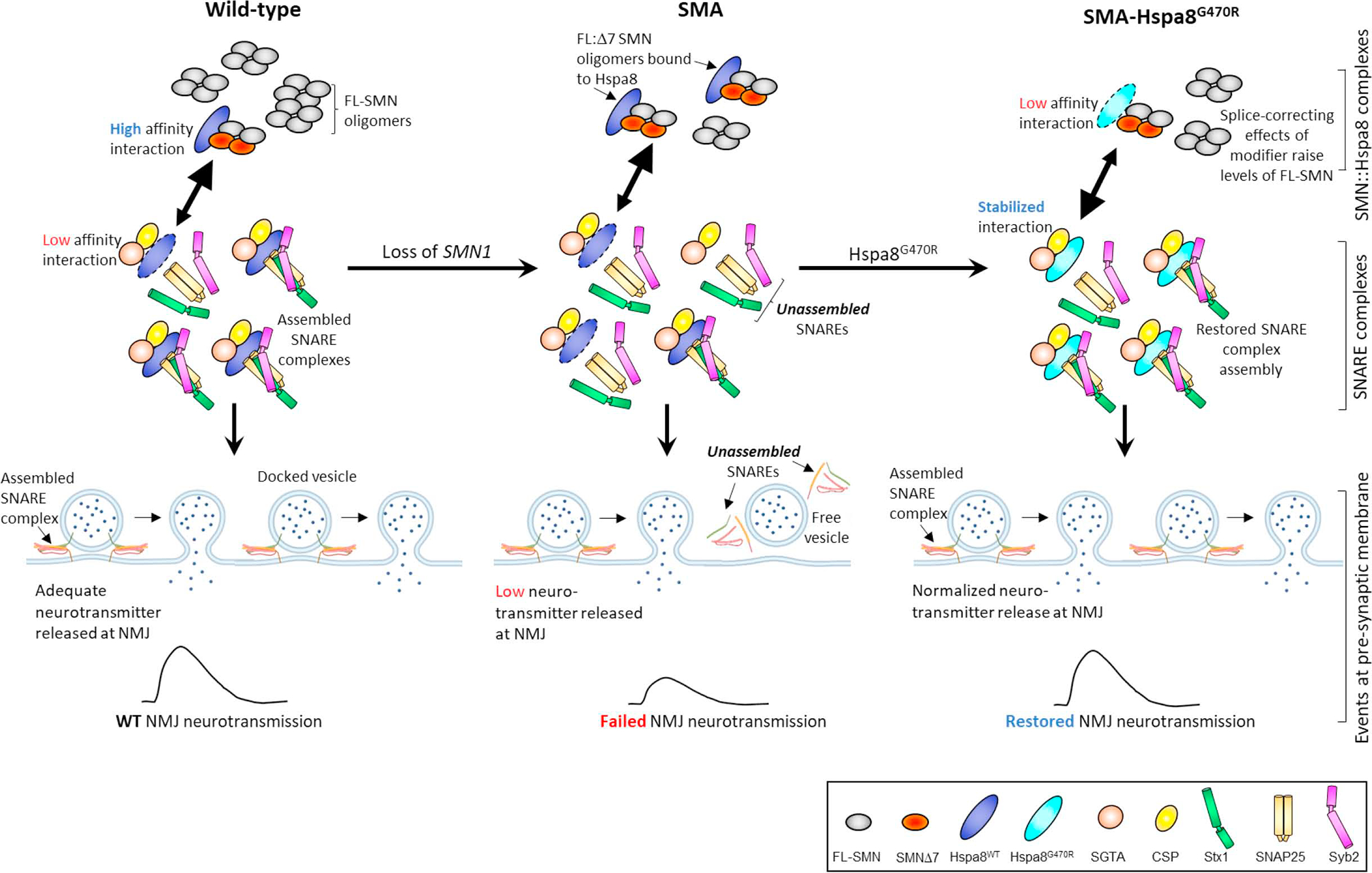
Depicted is a hypothetical model that encapsulates the relationship between SNARE complex assembly, SMN and the Hspa8G470R SMA modifier at NMJs. In the WT state, the preponderance of SMN exists as intact (FL), stable oligomeric complexes, SMNΔ7 isoforms being excluded from the complexes. Consequently, relatively little Hspa8 is required for SMN turnover in synapses. Hspa8 is therefore free to engage with its synaptic co-chaperones, CSPα and SGTA, notwithstanding a potentially weak affinity for these proteins. Still, this ensures proper SNARE complex assembly and efficient neurotransmission. In the absence of SMN1 (SMA), total FL-SMN levels fall and relative concentrations of oligomers of the protein containing the unstable SMNΔ7 species and proteins such as NF rise. Hspa8 is diverted away from the tripartite chaperone complex to effect turnover of dysregulated proteins such as the less stable FL-SMN:SMNΔ7 hybrid oligomeric complexes. Consequently, repeated cycles of SNARE complex assembly are disrupted. The G470R variant reverses this effect by stabilizing the interaction of Hspa8 with its co-chaperones and concomitantly weakening its association with SMN and perhaps other clients too. SNARE complex assembly is thus restored. A weak, SMN2 splice-switching property inherent in Hspa8G470R modestly raises levels of the intact SMN and combines with the effect of the variant on assembling SNAREs to potently suppress the SMA phenotype. Note: Larger arrowheads in double-headed arrows signify direction of preferred interaction.
