Fig. 1. Complete assembly of the Arabidopsis centromeres.
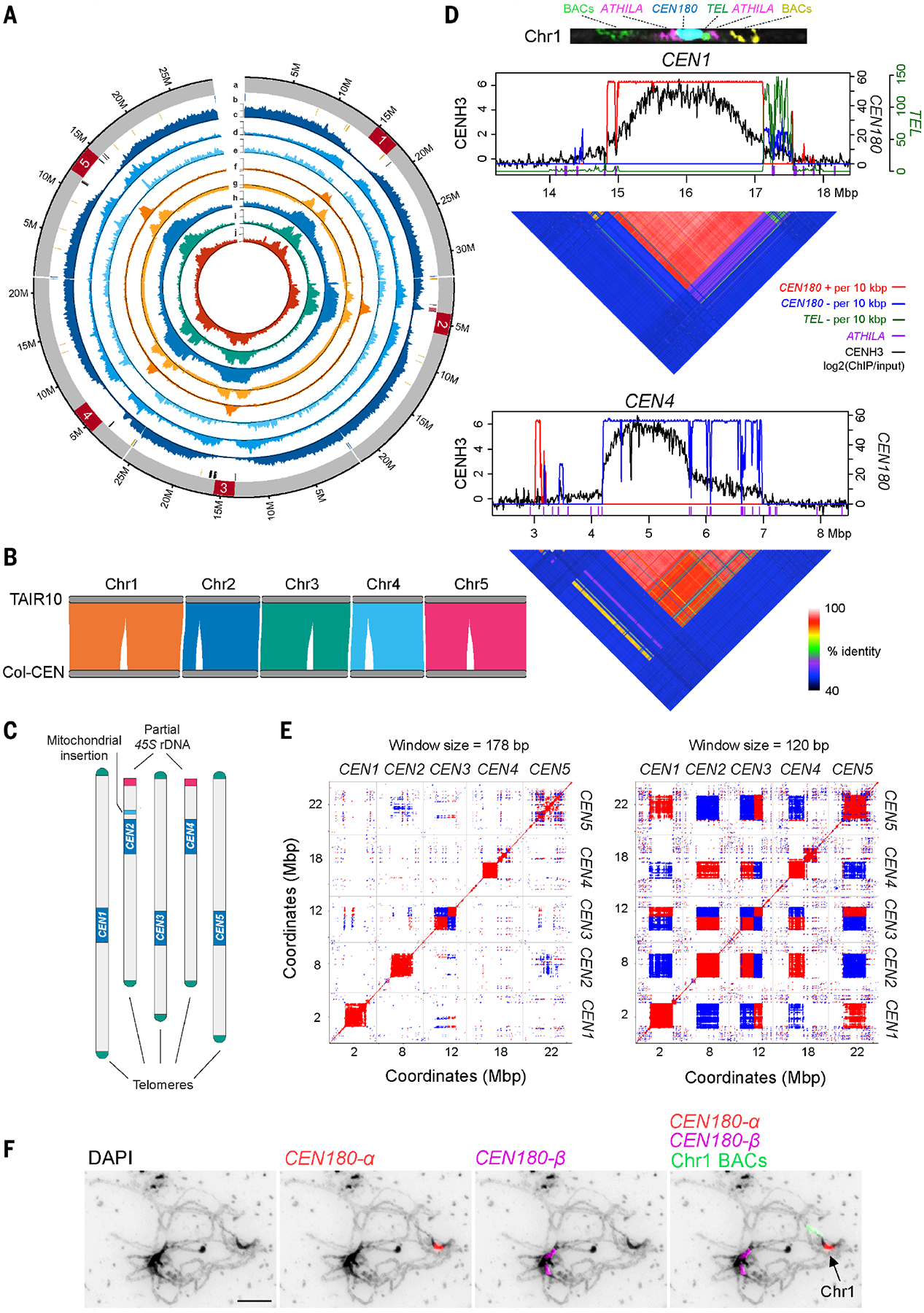
(A) Circos plot of the Col-CEN assembly. Quantitative tracks (labeled c to j) are aggregated in 100-kbp bins, and independent y-axis labels are given as (low value, mid value, high value, measurement unit) as follows: (a) chromosome with centromeres shown in red; (b) telomeres (blue), 45S rDNA (yellow), 5S rDNA (black), and the mitochondrial insertion (pink); (c) genes (0, 25, 51, gene number); (d) transposable elements (0, 84, 167, transposable element number); (e) Col×Ler F2 crossovers (0, 7, 14, crossover number); (f) CENH3 [−0.5, 0, 3, log2(ChIP/input)]; (g) H3K9me2 [−0.6, 0, 2, log2(ChIP/input)]; (h) CG methylation (0, 47, 95, %); (i) CHG methylation (0, 28, 56, %); and (j) CHH methylation (0, 7, 13, %). (B) Syntenic alignments between the TAIR10 and Col-CEN assemblies. (C) Col-CEN ideogram with annotated chromosome landmarks (not drawn to scale). (D) CENH3 log2(ChIP/input) (black) plotted over centromeres 1 and 4 (10). CEN180 per 10-kbp plotted for forward (red) or reverse (blue) strand orientations. ATHILA are indicated by purple x-axis ticks. Heatmaps show pairwise sequence identity between all nonoverlapping 5-kbp regions. A FISH-stained chromosome 1 at pachytene is shown at the top, probed with upper-arm BACs (green), ATHILA (purple), CEN180 (blue), the telomeric repeat (green), and bottom-arm BACs (yellow). (E) Dot plots comparing the five centromeres using a search window of 120 or 178 bp. Red and blue indicate forward- and reverse-strand similarity, respectively. (F) Pachytene-stage chromosomes stained with 4′,6-diamidino-2-phenylindole (DAPI) (black) and CEN180-α (red), CEN180-β (purple), and chromosome 1 BAC (green) FISH probes. The scale bar represents 10 μM.
