Fig. 3. Invasion of the Arabidopsis centromeres by ATHILA retrotransposons.
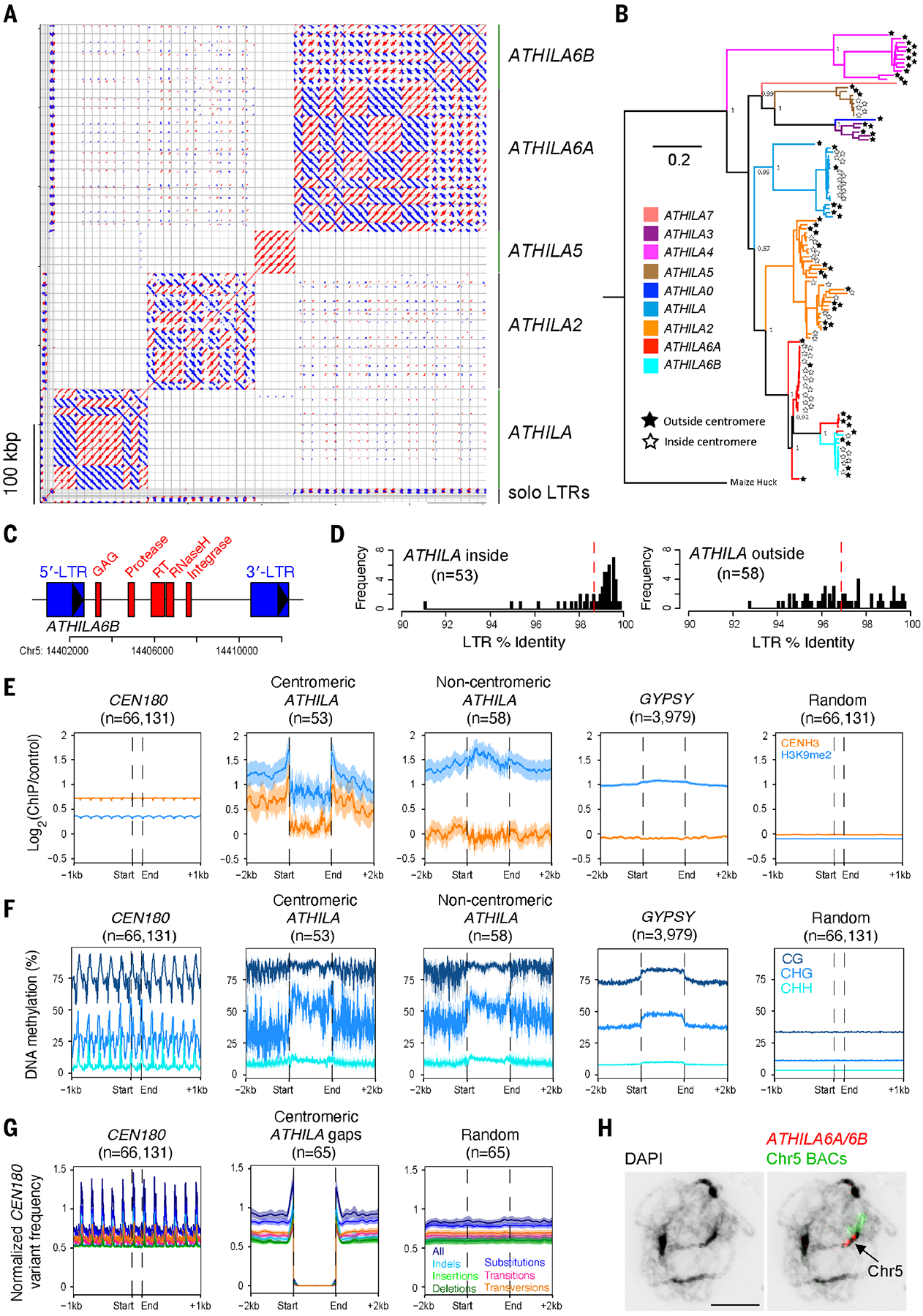
(A) Dot plot of centromeric ATHILA using a 50-bp search window. Red and blue indicate forward- and reverse-strand similarity, respectively. ATHILA subfamilies and solo LTRs are indicated. (B) Maximum likelihood phylogenetic tree of 111 intact ATHILA elements, color coded according to subfamily. Stars at the branch tips indicate ATHILA inside (white) or outside (black) the centromeres. (C) An annotated map of an ATHILA6B with LTRs (blue) and core protein domains (red) highlighted. (D) Histograms of LTR sequence identity for centromeric ATHILA elements (n = 53) compared with ATHILA outside of the centromeres (n = 58). Red dashed lines indicate mean values. (E) Metaprofiles of CENH3 (orange) and H3K9me2 (blue) ChIP-seq signals around CEN180 (n = 66,131), centromeric intact ATHILA (n = 53), ATHILA located outside the centromeres (n = 58), GYPSY retrotransposons (n = 3979), and random positions (n = 66,131). Shaded ribbons represent 95% confidence intervals for windowed mean values. (F) Same as for (E) but analyzing ONT-derived percentage of DNA methylation in CG (dark blue), CHG (blue), and CHH (light blue) contexts. (G) Meta-profiles of CEN180 sequence edits (insertions, deletions, and substitutions relative to the CEN180 consensus), normalized by CEN180 presence, in positions surrounding CEN180 gaps containing ATHILA (n = 65) or random positions (n = 65). All edits (dark blue), substitutions (blue), indels (light blue), insertions (light green), deletions (dark green), transitions (pink), and transversions (orange) are shown. Shaded ribbons represent 95% confidence intervals for windowed mean values. (H) Pachytene-stage chromosome spread stained with DAPI (black), an ATHILA6A/6B GAG FISH probe (red), and chromosome 5–specific BACs (green). The scale bar represents 10 μM.
