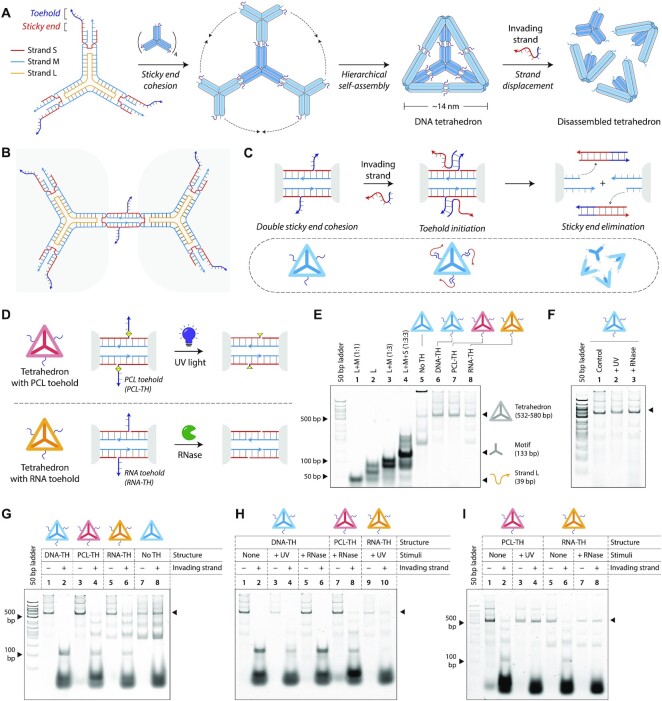
Figure 4.
Toehold clipping to control reconfiguration of a DNA tetrahedron. (A) Design and assembly of the 3-point-star motif into a DNA tetrahedron. The motif is constructed from three strands, a short (S), medium (M) and a long (L) strand (strand identities are indicated by different colors). An invading strand can bind to the toeholds on the DNA tetrahedron causing disassembly of the structure. (B) The 3-point-star motifs connect to each other via double cohesion. Beyond the sticky end region, the motif also contains a toehold. (C) An invading strand can bind to the toehold on strand S and remove it from the motif, this eliminating the sticky end cohesion and causing the tetrahedron to disassemble. (D) Schematic showing toehold clipping in the DNA tetrahedron using light or enzyme. (E) Non-denaturing gel showing stepwise assembly of the DNA tetrahedra. (F) Gel image showing DNA tetrahedra are unaffected on exposure to UV or on RNase A addition. (G) Tetrahedron disassembly by addition of invading strand. (H) Demonstration of stimuli specificity in DNA tetrahedron disassembly. (I) Toehold clipping by UV on PCL-TH tetrahedron and RNase A on RNA-TH tetrahedron prevents tetrahedron disassembly.