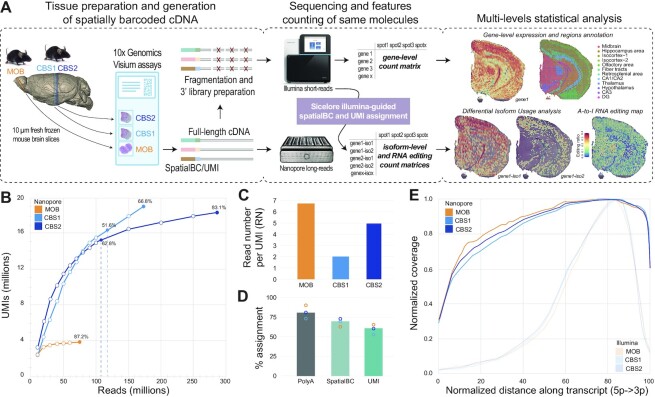
Figure 1.
SiT methodology and datasets. (A) Experimental and computational steps for SiT analysis. Right side shows unsupervised gene expression clustering and gene- (short-read) and isoform-level (SiT) expression of Snap25 in a mouse coronal brain section (CBS2). (B) Nanopore sequencing saturation curves for three Visium samples showing the number of UMIs observed as a function of the number of Nanopore reads. Labels indicate sequencing saturations obtained with all flow cells (CBS1, CBS2, MOB) and with just one latest generation Promethion flow cell per sample (vertical dotted lines, CBS1, CBS2). (C) Mean read number (RN) per molecule (UMI) observed for each of the three samples. (D) Percentage of assignment at each step of the workflow: Reads with polyA tail (PolyA) expressed as percentage of total reads; Assigned spatial barcode (SpatialBC) expressed as percentage of reads with PolyA tail found; UMI assigned reads (UMI) as percentage of reads with spatial barcode. Details about the spatial barcode/UMI assignment strategy are in (15). (E) Normalized transcript coverage plot for Nanopore and for Illumina sequencing.