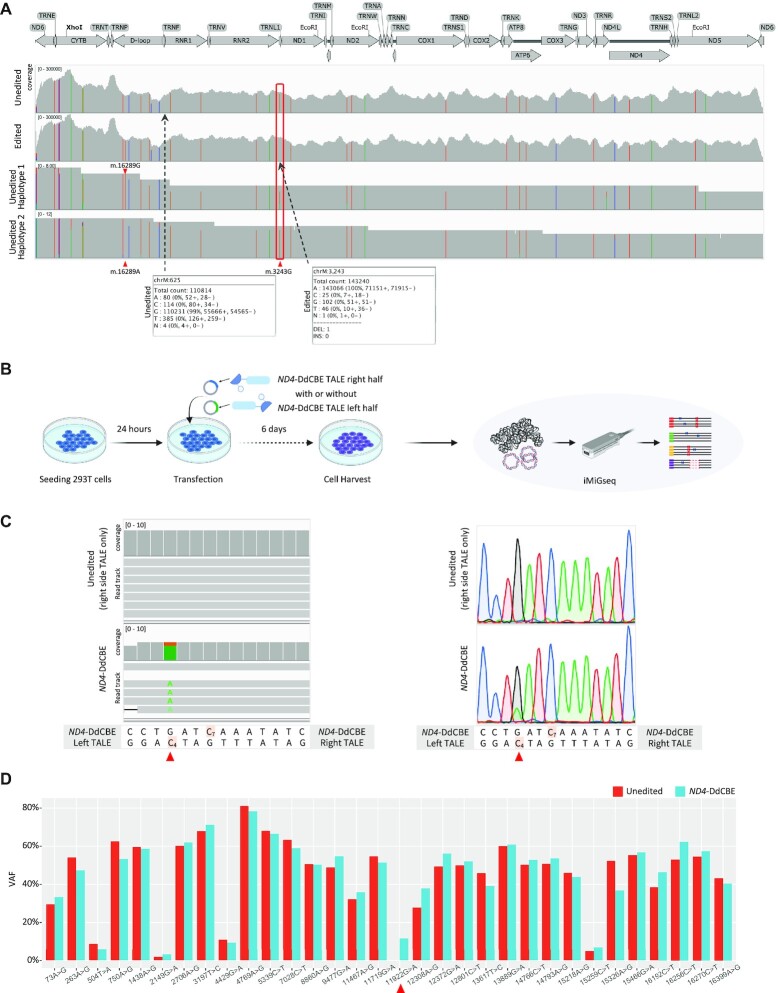
Figure 4.
Characterization of mitochondrial genome editing by mitoTALEN and DdCBE. (A) The read alignment of unedited (the top track) and mitoTALEN edited (the second track) cells by deep Illumina sequencing, and two haplotypes (the bottom two tracks) of unedited cells by iMiGseq illustrated by IGV plots. The mitoTALEN target position (m.3243) is labeled by red frame. Bulk Illumina sequencing confirms the complete elimination of the m.3243A > G mutation but fails to detect the rare m.625G > A SNV (VAF of 0.55%). The two haplotypes by iMiGseq show the predominant linkage between m.16289A and m.3243G, and the infrequent linkage of m.16289G and m.3243G (only one UMI group is shown for each haplotype). Bulk Illumina sequencing is unable to conduct haplotype analysis. (B) Schematic representation of mitoTALEN and DdCBE editing for iMiGseq. 293T cells were seeded in collagen-coated 48-well plate, cultured for 24 hours for transfection of TALEN/DdCBE left half and right half plasmids. The transfected cells were cultured for another 6 days before cell harvest, followed by DNA extraction and iMiGseq. (C) The m.11922G > A SNV (on the top H-strand, i.e. C4> T on the bottom L-strand) (red triangle) installed by ND4-DdCBE is detected by iMiGseq (left, one UMI group is shown for each genotype) and confirmed by Sanger sequencing (right). A diagram of the ND4-DdCBE left and right TALEs and the target spacing region is shown below the sequencing results. The cytosines that are within the ND4-DdCBE editing window are highlighted. Cells received only the right side ND4-DdCBE TALE are used as the unedited control. (D) The VAF changes of primary SNVs (VAF > 1%) before and after ND4-DdCBE editing. No significant VAF shift is observed.