Table 2.
CryoEM data collection, refinement, and validation statistics
| Data collection | Wild-type BrxL | E280Q-BrxL |
|---|---|---|
| EM equipment | Glacios | Krios |
| Voltage (kV) | 200 | 300 |
| Detector | Gatan K3 | Gatan K3 |
| Pixel size (Å/Pixel) | 0.561 | 0.5347 |
| Electron dose (e−/Å2) | 50 | 50 |
| Defocus range (μm) | 0.1–4.0 | 0.1–4.0 |
| Micrographs collected | 2982 | 7430 |
| Micrographs used | 2906 | 5298 |
| Reconstruction: | ||
| Software | cryoSPARC | cryoSPARC |
| Number of used particles | 22 162 | 234 715 |
| Symmetry | C7 | No symmetry |
| Resolution (Å) | 3.55 | 3.62 |
| Map sharpening B-factor (Å2) | 74.5 | 96.8 |
| Refinement: | ||
| PDB/EMD ID | 8EMC/EMD-28244 | 8EMH/EMD-28248 |
| Software | Phenix | Phenix |
| Cell dimensions | ||
| a = b = c (Å) | 454.72 | 384.98 |
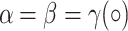
|
90 | 90 |
| Model composition: | ||
| Protein residues | 9328 | 8025 |
| Nucleotide | —- | 127 |
| Side chains assigned | 9328 | 8025 |
| MolProbity score: | 1.97 | 1.79 |
| Rms deviations: | ||
| Bonds length (Å) | 0.004 | 0.004 |
| Bonds angle (°) | 0.731 | 0.687 |
| Ramachandran plot statistics (%): | ||
| Favored | 95.01 | 97.08 |
| Allowed | 4.70 | 2.81 |
| Outlier | 0.11 | 0.29 |
