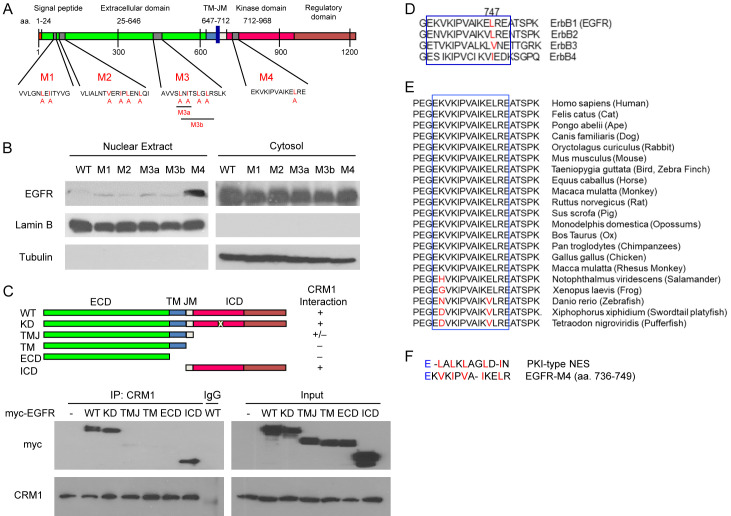
Figure 1.
Characterization of the putative NES on EGFR. (A) Diagram of the putative NES on EGFR in an in silico analysis. The human EGFR amino acid sequence was analyzed using the NESbase 1.0 database. The predicted NES sequences are referred to as M1, M2, M3, and M4. The red highlighted amino acid residue is replaced by alanine to generate NES mutants. (B) The putative NES regions were point-mutated with site-directed mutagenesis. The cytosolic and nuclear fractions were immunoblotted with anti-myc (EGFR). Lamin B and α-Tubulin were used as nuclear and cytosolic protein markers. (C) Myc-tagged deletion mutants of EGFR plasmids were transiently transfected into HEK293T cells. Whole cell lysates of the transfected cells were immunoprecipitated with anti-CRM1 antibodies. The precipitates were separated on SDS-polyacrylamide gel and immunoblotted with anti-myc and anti-CRM1. (D and E) The putative NES region sequence (M4) alignment of EGFR among four members of the ErbB family (D) and vertebrates (E). (F) Amino acid sequence similarity of EGFR and protein kinase inhibitor (PKI)-type NES. WT, wild-type; ECD, extracellular domain; ICD, intracellular domain; TM, transmembrane domain; JM, juxtamembrane domain; KD, kinase-dead.