Abstract
Glioblastoma is an aggressive brain tumor with a poor prognosis. Glioblastoma Stem Cells (GSC) are involved in glioblastoma resistance and relapse. Effective glioblastoma treatment must include GSC targeting strategy. Robust and well defined in vitroGSC models are required for new therapies evaluation. In this study, we extensively characterized 4 GSC models obtained by dedifferentiation of commercially available glioblastoma cell lines and compared them to 2 established patient derived GSC lines (Brain Tumor Initiating Cells). Dedifferentiated cells formed gliospheres, typical for GSC, with self-renewal ability. Gene expression and protein analysis revealed an increased expression of several stemness associated markers such as A2B5, integrin α6, Nestin, SOX2 and NANOG. Cells were oriented toward a mesenchymal GSC phenotype as shown by elevated levels of mesenchymal and EMT related markers (CD44, FN1, integrin α5). Dedifferentiated GSC were similar to BTIC in terms of size and heterogeneity. The characterization study also revealed that CXCR4 pathway was activated by dedifferentiation, emphasizing its role as a potential therapeutic target. The expression of resistance-associated markers and the phenotypic diversity of the 4 GSC models obtained by dedifferentiation make them relevant to challenge future GSC targeting therapies.
Keywords: Glioblastoma, glioblastoma stem cells, brain tumor initiating cells, gliospheres, in vitromodel, dedifferentiation
Introduction
Glioblastoma is the most frequent and aggressive primary brain tumor. Low treatment efficiency and constant relapses are responsible for a poor prognosis, with a median overall survival of 15 months [1]. Microenvironment interactions are involved in disease progression and therapeutic resistance through several mechanisms including glioblastoma stem cells (GSC) development and maintenance [2]. The presence of GSC within the tumor is associated with relapse and resistance to treatment [3]. Indeed, GSC are able to self-renew and can differentiate into glioblastoma cell populations depending on their phenotype [4]. GSC can be mesenchymal or proneural and each phenotype being associated to different levels of aggressivity and therapeutic sensitivity [5]. Mesenchymal GSC are more susceptible to be involved in relapse due to their invasive and resistant profile. In response to radiation and cytotoxic treatments, proneural GSC can shift toward a mesenchymal phenotype through epithelial-mesenchymal transition (EMT) [6-8]. To identify new therapeutic targets, it is crucial to better understand cell and molecular mechanisms involved in GSC development and therapeutic escape.
To study GSC, two main in vitro models can be used: patient derived cells or cells dedifferentiated from glioblatoma cell lines. GSC from both origins typically grow as gliospheres, 3D cell clusters in suspension. The ability to form gliospheres in vitro is a common feature to validate the stemness of the obtained models. Patient derived GSC are usually purified from tumor sample, based on the expression of one or two enrichment markers such as CD133 and CD15 [9,10]. However, CD133 significance as a single GSC marker is controversial. Indeed, proneural GSC are known to express CD133 but mesenchymal GSC are CD133 negative, while they are more resistant and aggressive [11]. Other studies used CD44 and CD133 to isolate mesenchymal and proneural cells from the same sample [12] but some GSC can have a complex phenotype, expressing both markers [11]. Considering GSC heterogeneity, markers-based isolation induces a loss of diversity compared to the initial tumor. Another protocol is based on cultivating tumor samples in a GSC promoting medium [13]. Working with patient derived cells is advantageous because cells are closer to their in vivo characteristics, but requires accessibility to patient samples. To avoid in vitro drift, GSC can be kept as xenograft, being harvested only before experiment [10]. Such models are complex and involve ethical considerations and important financial costs, preventing large scale studies and availability for multiple laboratories. Dedifferentiated gliospheres are easy to produce from commercially available cell lines and have been used as GSC models in several studies [14-16]. An advantage of this model is the possibility to compare GSC and their differentiated counterparts to reveal stemness specific characteristics and pathways involved in dedifferentiation for target identification.
GSC models are usually validated and characterized using some of the most known markers (CD133, SOX2, CD15 (SSEA-1), A2B5, CD44) and cellular characteristics (sphere-forming ability, self-renewal) [12,14,17-19]. GSC enrichment based on markers expression is limited since the cells can express a multitude of markers involved at varying degrees in stemness, growth, invasion and resistance. Before performing therapy evaluation, GSC should be extensively characterized in a more extensive manner to ensure stemness and phenotype.
The objective of this study was to establish robust, accessible and well-characterized GSC models that can be used to identify therapeutic targets. We performed an extensive analysis of 4 GSC models obtained by dedifferentiation of commercially available GBM cell lines and compared them to 2 patient-derived GSC lines. We demonstrate that dedifferentiated GSC show similarities to patient derived stem cells and have a heterogeneous, complex profile, susceptible to challenge therapy evaluation. Based on these models characterization, we provide support for therapeutic targets among CXCR4 pathway.
Material and methods
Cell culture
Glioblastoma cell lines U-87MG, U-118MG, U-251MG and T98-G were provided by ATCC. Adherent cells were maintained in DMEM high glucose (Sigma-Aldrich®) supplemented with 10% FBS (Biowest) and 1% penicillin/streptomycin solution (Gibco®) at 37°C, 5% CO2. Cells were passaged using trypsin 0.25% (Sigma-Aldrich®) every week and kept in culture for 6 consecutive weeks maximum. Two GSC lines, BTIC 25m and 12m, were obtained from Dr H. Artee Luchman and from Pr. Samuel Weiss (Hotchkiss Brain Institute). BTIC were cultivated using NeuroCult™ NS-A Proliferation Kit Human (Stemcell®). Gliospheres were dissociated using accutase (Sigma-Aldrich®) and diluted every week, for less than 6 consecutive weeks.
Neuroblastoma cell lines (SH-SY5Y and NGP) as well as cervix adenocarcinoma cell line (HeLa) were purchased from ATCC. Normal human astrocytes (NHA) were acquired from Lonza Bioscience®. Cells were cultivated in similar conditions as glioblastoma cell lines.
Dedifferentiation protocol
Adherent cells from U-87MG, U-118MG, U-251MG and T98-G cell lines were harvested at 70-80% confluence, washed with DPBS (Gibco®) and resuspended in dedifferentiation medium and incubated at 37°C, 5% CO2. Dedifferentiation medium, adapted from [20], was composed of DMEM/F-12 (Gibco®) supplemented with 1% N-2 (Gibco®), 1% B-27 (Gibco®), 20 ng/mL hEGF (human Epidermal Growth Factor, Miltenyi®), 20 ng/mL bFGF-2 (basic Fibroblast Growth Factor, Miltenyi®) and 1% penicillin/streptomycin. After 24 h, cells were mechanically resuspended and fresh medium was added. Gliospheres were harvested when they reached their maximum size before impact on cell viability (to avoid necrotic core): at 48 h for U-251MG and T98-G cell lines, at 96 h for U-118MG and U-87MG cell lines. A medium addition was performed at 72 h for 96 h incubation time.
Morphological characterization
Gliospheres pictures were recorded using an inverted microscope (Zeiss®, Primovert) when they reached their maximum growth before viability decreases. Gliosphere areas were measured with ImageJ software thanks to the help of J. Chaigneau (image analysis engineer). Briefly, images were smoothed, a threshold was set to remove background before analysis by particle detection ImageJ plugin. Particle area was converted from pixel2 to µm2 using picture scale. Such analysis was performed on pictures from 3 independent experiments, giving areas measurement for at least 100 gliospheres.
Self-renewal assessment
Gliospheres were dissociated using accutase and decreasing cell quantity (400 to 1) was seeded in 96-well plates containing 200 µL dedifferentiation medium. Each condition was replicated in 10 wells. To quantify gliosphere forming yield, gliosphere number in 5 wells containing 100 cells each was recorded. Data were gathered and analyzed from 3 independent experiments.
RT-qPCR
The expression of 31 GSC-associated genes was measured by RT-qPCR to identify key markers for further studies at the protein level (see Table S1 for markers information). Adherent cells and gliospheres were harvested and washed with cold DPBS before storage as dry pellet at -80°C. RNA extraction and RT-qPCR were carried out by J. Cayon and L. Bonneau PACeM platform (Plateforme d’Analyse Cellulaire et Moléculaire, SFR ICAT 4208, Angers, France). Primer sequences are property of PACeM platform and are available on request. Total mRNAs were extracted with RNeasy Micro-kit (Qiagen) according to the manufacturer’s instructions. Quantification was performed using NanoDrop2000 (ThermoFisher Scientific®). cDNAs were generated using the Reverse Transcriptase SuperScript II® kit (Invitrogen®) and purified using QIAquick® PCR purification kit (Qiagen®) according to the manufacturer’s instructions. Specific primers were designed on Primer Blast website. qPCR was performed using LightCycler 480 II (Roche) and SYBR Green as fluorescent probe. Cq were converted to relative mRNA expression according to Vandesompele method [21], with two reference genes: GAPDH and HPRT1 and the most expressed gene as internal calibrator. To easily compare genes expression between adherent and dedifferentiated cells, fold expression was calculated following Livak method [22] with mean reference genes Ct and adherent cells as control conditions for each cell line. Fold expression <0.5 indicates a decreased expression in gliospheres while a fold expression >2 shows overexpression. Heatmaps representing fold expression and relative expression were created with the R package ComplexHeatmap version 2.12.1 [23,24].
Flow cytometry
For stem cell phenotype evaluation, adherent cells and gliospheres were stained using a panel of antibodies against 10 known GSC markers (CD44, CD133, A2B5, CD90, integrin α-5, integrin α-6, CXCR4, CD74, CD15 and CD36) for flow cytometry analysis (see Table S1 for markers information). Antibodies and corresponding isotype controls were purchased from Miltenyi® (see Table S2 for complete panel references). Adherent cells and gliospheres were dissociated using accutase, washed with corresponding complete medium and 250 K cells were resuspended in 50 µL DPBS containing 0.5% BSA and 2 mM EDTA (FC buffer). Cells were stained 15 min at 4°C with antibodies at 1:50 dilution, except for anti-CD74 antibody, at 1:11 dilution. Cells were washed twice with FC buffer. Ten thousand events were recorded using CytoFLEX® flow cytometer equipped with CytExpert® software (material from PACeM platform). Compensation matrix was automatically established with single-stained samples, manually corrected, and was then applied on registered data. Cytometry measurements were processed with FlowJo® V10 software. Positive population gating used isotype controls as references, giving the percentage of positive cells in each sample. Mean fluorescence intensity (MFI) was calculated for each sample and was normalized on corresponding isotype control MFI. The MFI ratio obtained enables comparison between samples.
Immunocytochemistry
To complete GSC marker analysis, the expression of 3 proteins was evaluated in gliospheres and adherent cells by immunocytochemistry (see Table S1 for markers information). Olig2 transcription factor and cytoskeleton components (Nestin and GFAP) were analyzed. Adherent cells were seeded on glass coverslips during 48 h at 37°C, 5% CO2. Gliospheres were seeded on CellTak® (Corning®) coated coverslip (3.5 µg/cm2) during 45 min at 37°C, 5% CO2. Cells were washed three times with DPBS 1% BSA between each protocol step. Fixation was performed in 4% paraformaldehyde (Euromedex®) during 15 min for gliospheres and 10 min for adherent cells. Samples were incubated 1 h in DPBS 1% BSA containing 0.1% Triton X-100 before overnight incubation at 4°C in primary antibody solutions (see Table S3 for antibodies information). Triton X-100 concentration was increased to 0.3% for intranuclear staining (Olig2). Primary antibodies were revealed using AF488 or AF568 anti-rabbit IgG antibodies (ThermoFisher Scientific®) at 1:250 for 2 h. Finally, nuclei were stained with 3 µM DAPI (Sigma-Aldrich®) for 15 min. Coverslips were mounted on SuperFrost® slides (ThermoFisher Scientific®) using ProLong Gold antifade solution (ThermoFisher Scientific®). Slides were observed with a confocal microscope (Leica TCS SP8 from Leica Biosystems®) with the help of R. Perrot from SCIAM platform (Service Commun d’Imageries et d’Analyses Microscopiques, SFR ICAT 4208, Angers, France).
Statistical analysis
Statistical analyses were performed with GraphPad® Prism® 8.3 software (GraphPad® Software, LLC). All experiments, except immunocytochemistry, were repeated at least 3 independent times. Means comparisons were performed with Mann-Whitney test, differences were considered significant at P<0.05.
Results
Dedifferentiated gliospheres are morphologically similar to patient derived GSC
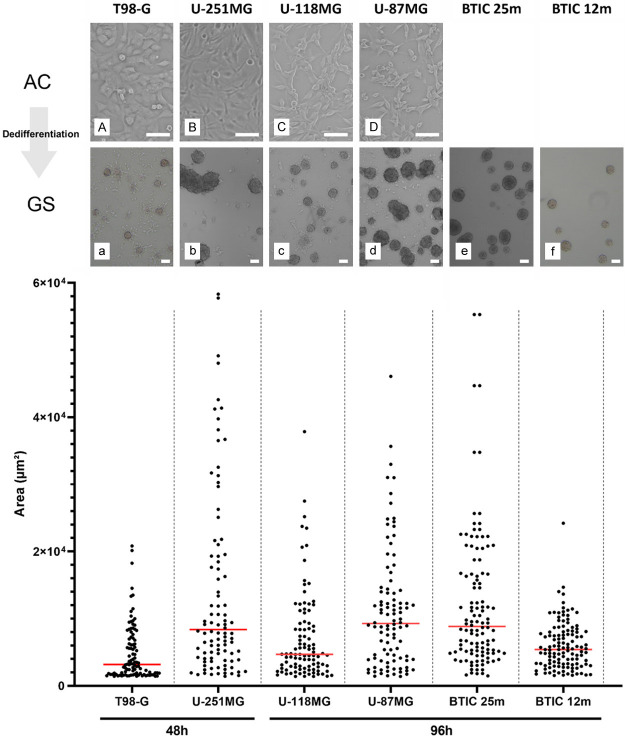
We first dedifferentiated 4 GBM cell lines (U-87MG, U-251MG, U-118MG and T-98G) using a standardized and accessible protocol to obtain GSC and compared their morphology to 2 patient-derived GSC lines (BTIC 12m and 25m) as standards. As for cancer and neural stem cells, glioblastoma stem cells cultivated in vitro are known to form 3D cell clusters in suspension named “gliospheres” (GS). As illustrated by Figure 1 (top), when cultivated in dedifferentiation medium, adherent GBM cell lines rapidly grown as gliospheres, closely alike BTIC. This morphological change was specific to glioblastoma cells since it was not observed in normal astrocytes (NHA) nor neuroblastoma (SH-SY5Y and NGP) or cervix adenocarcinoma (HeLa) cell lines in the same culture conditions (Figure S1). Their morphology varied across cell lines, some gliospheres being partly adherent and smaller (T-98G, U-118MG, BTIC 12m) with others being completely floating (U-87MG, U-251MG, BTIC 25m) and sometimes forming gliosphere aggregates (U-251MG). To perform quantified comparison between cell lines, gliosphere areas were measured from pictures taken with an inverted microscope. Areas displayed in Figure 1 illustrate size heterogeneity within the same cell line, which corroborates picture observations. Despite variations between cell lines distribution, areas were similar. Particularly, U-251MG, U-87MG and BTIC 25m had similar medians while U-118MG were closer to BTIC 12m, as previously noted on pictures. However, important differences in behavior could be noted for T98-G and U-251MG. T98-G gliospheres were smaller, had a shorter distribution and their size did not increase after 48 h. U-251MG gliospheres grew rapidly, reaching large sizes in only 48 h of dedifferentiation. Moreover, the cells tended to stick to each other, forming gliosphere aggregates. All these results suggest that gliosphere morphology is cell line dependent, emphasizing the need for multiple models to represent GSC heterogeneity.
Figure 1.
Dedifferentiated gliospheres are similar to BTIC for appearance, size and heterogeneity. Microscope observations before/after dedifferentiation of T98-G (A, a), U-251MG (B, b), U-118MG (C, c), U-87MG (D, d), BTIC 25m (e) and 12m (f). Capital letters are used for adherent controls (AC) and lowercase letter for gliospheres (GS). Pictures were taken when gliospheres reached their maximum viable growth size. Scale is 10 µm. Corresponding gliospheres sizes are shown below. Areas were measured using ImageJ software for 100 gliospheres obtained from 3 independent experiments. Medians are represented in red.
Dedifferentiated gliospheres have GSC-like self-renewal abilities
GSC are characterized by their ability to self-renew after dissociation. GSC should maintain themselves in a GSC promoting medium containing growth factors. As gliospheres are composed of GSC and more differentiated cells, dissociation enables to separate cell populations and observe cell types. All our dedifferentiated cell lines were able to form gliospheres after dissociation, at different yields (Figure 2A). U-87MG and U-118MG cells generated gliospheres with similar size when compared to original gliospheres, showing high growth capacity (Figure 2B). However, T98-G and U-251MG gliospheres were much smaller (Figure 2B), with a low growth rate. Such results may suggest a quiescent GSC profile. Particularly, in T-98G and U-251MG cell lines, adherent cells were observed, indicating heterogeneity in the original gliospheres. Overall, dedifferentiated U-118MG and U-87MG seem closer to BTIC 25m in terms of yield and growth while T-98G and U-251MG resemble BTIC 12m.
Figure 2.
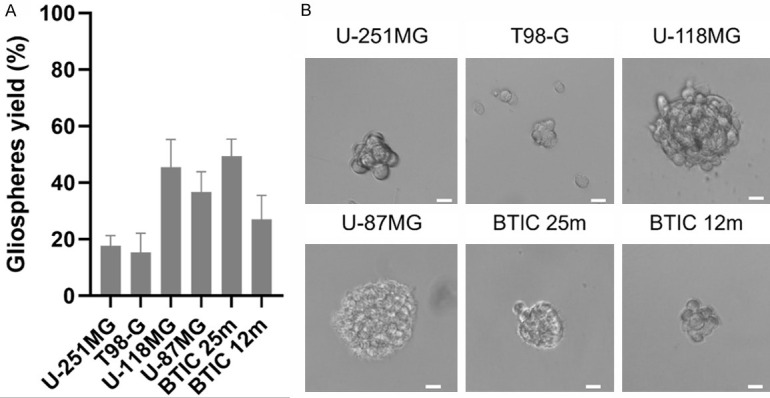
Dedifferentiated cells self-renew and reform gliospheres after dissociation. A. Yield in gliospheres calculated from their number in wells seeded with 100 cells. Mean represented with standard deviation from 15 measurements across 3 independent experiments. B. Examples of spheres formed in wells with 100 cells. Scale is 20 µm.
Dedifferentiation conditions induce a gene expression shift toward stem-cell, EMT and CXCR4 pathways
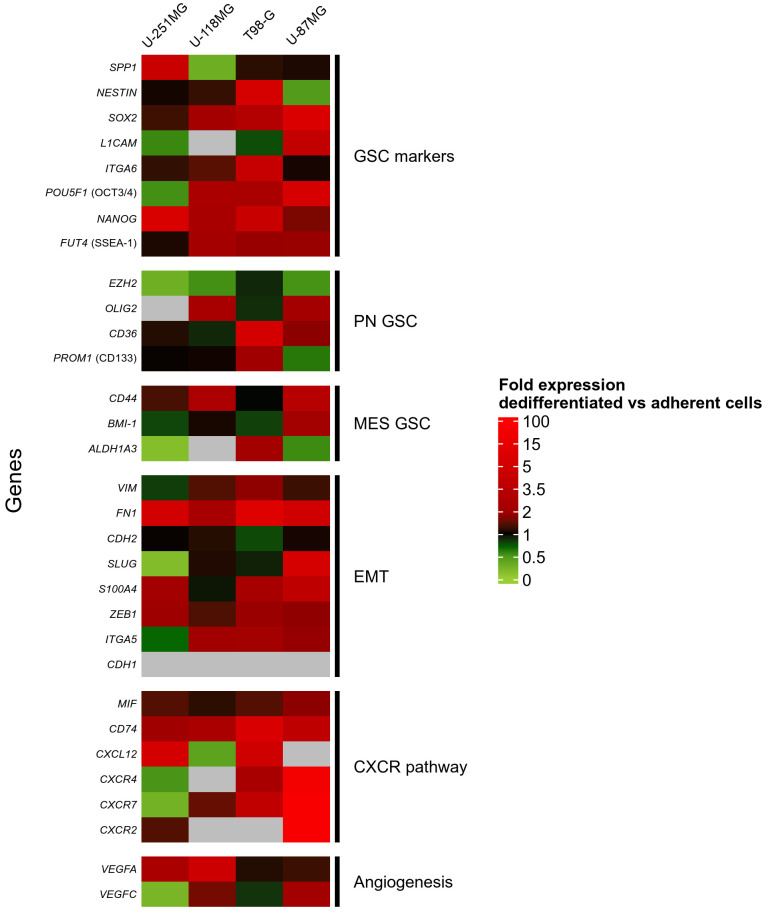
To characterize our models, we compared 31 genes expression levels between dedifferentiated gliospheres (GS), corresponding adherent counterparts (AC) and BTIC. Genes were chosen according to a literature review on GSC and glioblastoma prognostic markers (See Table S1 for complete markers information). Fifteen genes are associated with stem-cell maintenance and/or commonly used as GSC markers. Among them, 4 are known to be specific for the proneural (PN) phenotype (such as PROM1 or CD133) and 3 for the mesenchymal (MES) phenotype (such as CD44). 8 genes associated with Epithelial-Mesenchymal Transition (EMT) were also included in the study as they can be expressed in GSC. These genes can be associated with therapeutic resistance through PN to MES phenotype conversion (such as SLUG), sometimes even being GBM prognostic markers (such as CDH2). Associated with EMT, CXCR4-related pathway has been identified as a key actor of GSC maintenance and therapeutic resistance [25]. The 4 receptors CXCR4, CXCR2, CXCR7 and CD74 and their 2 ligands MIF and CXCL12 (also known as SDF-1) were included in our study. Finally, VEGFA and VEGFC were studied for their implication in angiogenesis and EMT.
The Figure 3 shows the global mRNA expression of the analyzed genes among all samples according to Vandesompele analysis method (see Figure S2 for complete heatmap including adherent cells). Such method allows comparison between genes and between cell lines. Results are reported on a heatmap with blue to red colors indicating low to high expression level. Different mRNA expression patterns could be seen between GSC models, even between BTIC 25m and 12m. A common tendency was nevertheless noticed for some well-expressed genes (FN1, EZH2, CD44, BMI1, VIM, CDH2) as well as for low/not expressed ones (POU5F1 (OCT3/4), NANOG, FUT4 (SSEA-1), CD36, PROM1 (CD133), CDH1, CXCR2). Some genes appeared major, reaching high expression levels, but only for some GSC models (SPP1, NES, SOX2, SLUG, S100A4, ZEB1, MIF). All the 4 generated models expressed a wide variety of GSC associated genes and related pathways, underlining their GSC profile. According to their global expression profiles, the models seemed to be oriented toward a mesenchymal GSC phenotype: low expression of PN markers (PROM1 (CD133), CD36) while MES-specific and EMT-related genes were generally highly expressed (CD44, BMI1, VIM, CDH2).
Figure 3.
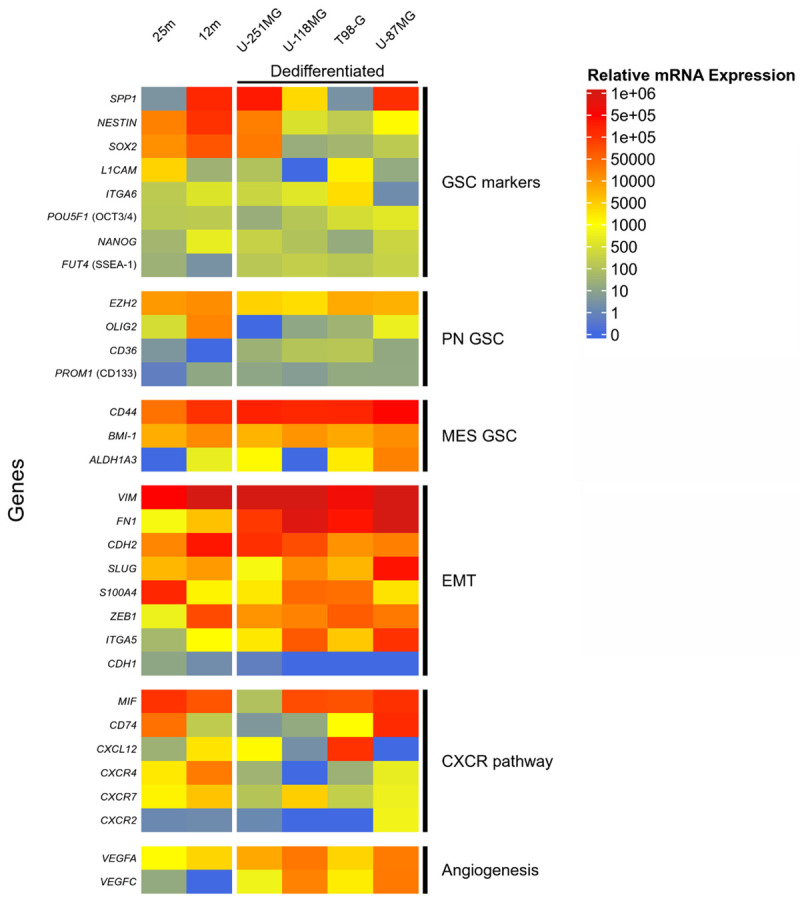
Transcriptional expression supports a mesenchymal GSC profile in gliospheres associated with high expression of some EMT, angiogenesis and CXCR associated genes. RT-qPCR analysis of gene expression in U-251MG, U-118MG, T98-G and U-87MG dedifferentiated gliospheres and in BTIC 25m and 12m. Relative mRNA expression was calculated and normalized according to Vandesompele method with two control genes (GAPDH and HPRT1) and an internal calibrator. Expression levels are colored from blue to red with increasing values. Genes are sorted by mean expression level in each group. PN: Proneural, MES: Mesenchymal, EMT: Epithelial-Mesenchymal Transition.
To estimate the transcriptional impact of the dedifferentiation, fold expression levels in gliospheres vs adherent cells were analyzed (Figure 4, see Figure S2 for global mRNA expression). Despite some heterogeneity between cell lines, GSC-associated gene expression was clearly increased after the dedifferentiation process. Particularly, we observed an enhancement of SOX2, FUT4 (SSEA-1), POU5F1 (OCT3/4) and NANOG, transcriptional factors essential for stem cell phenotype acquisition or maintenance. Mesenchymal orientation was not only kept but emphasized by dedifferentiation as shown by the increase in CD44 and EMT-related gene expression. However, even if CD133 remained weakly expressed, its expression was conserved or increased in 3 dedifferentiated models, further supporting a GSC phenotype acquisition. Interestingly, several genes of the CXCR4-associated pathways were more expressed after dedifferentiation, especially in U-87MG and T-98G, bringing new evidence for the importance of this pathway in GSC phenotype. VEGF pathway, moderately expressed and stimulated by dedifferentiation conditions in some cell lines could be also identified as a marker for EMT and aggressiveness of the obtained GSC.
Figure 4.
Dedifferentiated GSC overexpress stem-cell transcriptional markers, EMT-related genes and CXCR4 pathway. RT-qPCR analysis of gene expression in U-251MG, U-118MG, T98-G and U-87MG was used to calculate fold expression in dedifferentiated gliospheres with adherent cells as reference. A fold expression >2 (red) means an overexpression in GS whereas a fold expression <0.5 (green) means a decreased expression in GS. Grey color stands for conditions with undetectable expression in gliospheres and in adherent cells. N: Proneural, MES: Mesenchymal, EMT: Epithelial-Mesenchymal Transition.
All these data support the acquisition of a GSC phenotype after dedifferentiation, with a gene expression profile comparable to the one of patient-derived stem cells such as BTICs. The obtained cells appear to belong to a more resistant phenotype, mainly mesenchymal, as shown by the high expression of numerous EMT-related genes as well as CXCR4 and VEGF pathways.
Flow cytometry highlights important GSC markers in dedifferentiated gliospheres
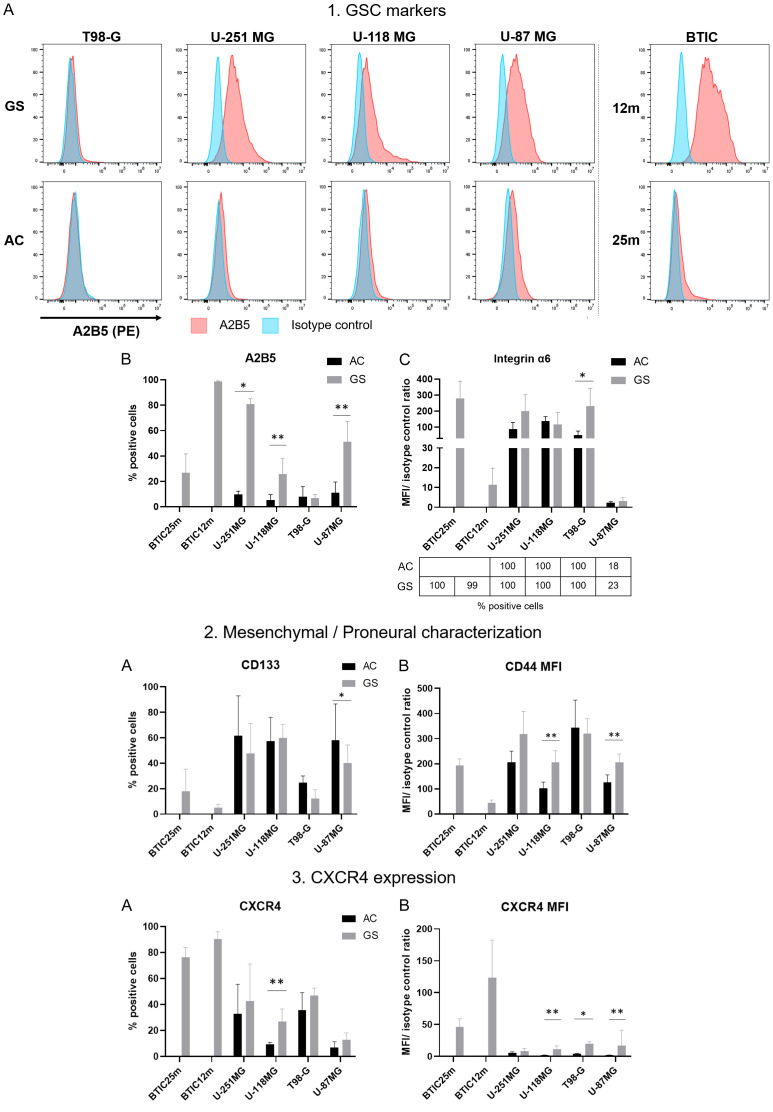
To confirm GSC markers at the protein level, the expression of 10 surface markers (A2B5, Integrin α5 and 6, CD133, CD44, CD90, CD36, CD15 (SSEA-1), CXCR4 and CD74) was analyzed by flow cytometry in BTIC, dedifferentiated gliospheres and corresponding adherent cells (See Table S1 for complete markers information).
A2B5, Integrin α6, CD15, CD90 and CD36 are GSC markers, the first three being used for GSC enrichment. Interestingly, A2B5 expression was negative in adherent cells and was detectable only after dedifferentiation in U-118MG, U-251MG and U-87MG cells (Figure 5-1A, 5-1B). Emphasizing once more GSC variability, this marker was not expressed in either T98-G gliospheres and in BTIC 25m while expressed in BTIC 12m. Similarly, integrin α-6 (or CD49f, coded by ITGA6), was highly expressed in all studied models (except in U-87MG). Its signal intensity was increased by dedifferentiation in U-251MG and T98-G (Figure 5-1C). These results support stemness characteristics acquisition induced by dedifferentiation. CD90 was not ubiquitous and did not seem impacted by dedifferentiation. Indeed, CD90 was highly expressed (100% positive cells, high MFI) in U-118MG, U-251MG and T-98G, with little change in GS (Figure S3). In BTIC, CD90 was only expressed in 57% of 12m cells. Our data is consistent with the literature as CD90 is not necessary for stem cell maintenance [26-28]. Finally, CD36 and CD15 were not revealed in any of our sample (data not shown), despite previous studies on U-251MG and U-87MG gliospheres showing CD36 expression in both cell lines and low CD15 expression in U-251MG [18].
Figure 5.
Change in membrane markers expression supports acquisition of GSC with a mainly mesenchymal phenotype in dedifferentiated gliospheres models. Flow cytometry analysis of GSC markers (A2B5 and integrin α-6), mesenchymal/proneural markers and CXCR4 in U-87MG, U-118MG, U-251MG, T98-G and BTIC 25m and 12m. Adherent cells (AC) were used as control for dedifferentiated gliospheres (GS). A2B5 staining results are shown as typical flow cytometry histograms (1A) and corresponding positive population percentage (1B). Integrin α6 analysis is depicted as mean fluorescence ratio with corresponding positive cells percentage in the table below (1C). Percentage of positive cells are represented for CD133 (2A) and CXCR4 (3A). Mean fluorescence ratios are shown for CD44 (2B) and CXCR4 (3B). Mean fluorescence ratio is calculated as mean fluorescence intensity (MFI) of stained sample on corresponding isotype control MFI. Statistical significance was calculated using Mann-Whitney test on 4 independent experiments, except for U-87MG (n=6) and U-118MG (n=5). *p-value <0.05, **p-value <0.001.
CD44 and CD133 markers analysis was used to confirm the GSC phenotype. In line with RT-qPCR data, CD44 was expressed in all studied cells and its expression was significantly increased after dedifferentiation in U-118MG, U-251MG and U-87MG (Figure 5-2B). However, despite its low transcriptional expression, CD133 was detectable in all studied cell lines except BTIC 12m (Figure 5-2A). Low CD133 expression in BTIC is consistent with previous results [29]. Dedifferentiation induced a decrease in CD133 population in U-251MG, T-98G and U-87MG suggesting a proneural phenotype loss. Moreover, integrin α5, an EMT marker, was highly expressed in all cell lines, including BTIC at a lower level and slightly increased with dedifferentiation (Figure S3). Altogether, these results suggest that dedifferentiation directs cells toward a more aggressive mesenchymal profile, supporting CD44 and EMT-associated marker expression instead of CD133.
As the CXCR4 pathway seemed activated in our RT-qPCR data, we analyzed the expression of CXCR4 and CD74 receptors by flow cytometry. However, despite high transcriptional expression of its main ligand MIF, CD74 was expressed only in 30% of BTIC 25m and U-87MG gliospheres, with a slight increase compared to adherent cells (22%) (Figure S3). CXCR4 protein expression was higher in BTIC and was increased after dedifferentiation in the 4 cell lines, supporting both the stemness of our models and the significance of CXCR4 pathway in GSC (Figure 5-3A and 5-3B). Interestingly, in U-118MG gliospheres, the CXCR4 positive population was higher in A2B5 positive cells, showing an association between these markers (25% vs 52%) (Figure S4).
Immunocytochemistry reveals the expression of Nestin and GFAP in GSC
To confirm the dedifferentiation state of gliospheres, immunocytochemistry was used to characterize the expression of cytoskeleton components being GSC (Nestin) or glial differentiation (GFAP) markers. GFAP was present in U-251MG, U-118MG and U-87MG and BTIC (Figure 6). GFAP expression could be surprising, as it was previously considered as a mature astrocytic marker and thus, a BTIC differentiation marker [30]. However, it can also be expressed in neural stem cells and in glial cells at various differentiation state [31]. GFAP expression, albeit controversial, supports the glial profile of the 6 GSC models. The GSC marker Nestin was expressed in all studied cell lines, in a particularly strong manner in gliospheres (Figure 6). Its expression appeared increased by the dedifferentiation process, emphasizing Nestin importance in gliosphere structure while supporting acquisition of GSC characteristics.
Figure 6.
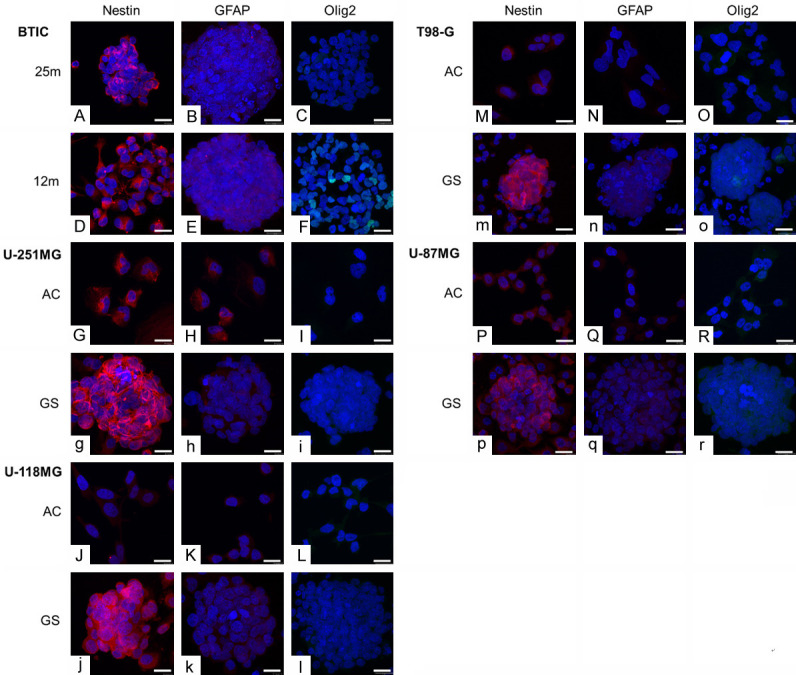
Expression of stem cells and glial marker proteins in adherent cells and in gliospheres. Typical fluorescent confocal photographs of cells immunostained to reveal Olig2 (AF488, green), Nestin (AF568, red), GFAP (AF568, red) and nuclei (DAPI, blue). Scale is 20 µm. Pictures represents BTIC 25m (A-C) and 12m (D-F), U-251MG (G-I), U-118MG (J-L), T98-G (M-O) and U-87MG (P-R), Capital letters stands for adherent cells (AC) and lowercase letters for gliospheres (GS).
Finally, the presence of Olig2 transcription factor was screened. Olig2 signal was very weak except in BTIC 12m (Figure 6), consistently with the RT-qPCR results.
Discussion
Pharmacological research on glioblastoma requires glioblastoma stem cell models for the identification and evaluation of targeted treatments. Each model has its own limitations, such as accessibility, lack of heterogeneity, low reproducibility, drifting due to long term culture or differences from in vivocharacteristics. Among available models, dedifferentiation of glioblastoma cells is frequently used as an easy way to obtain GSC, but these models are often poorly characterized. In this study, we characterize GSC obtained from 4 commercially available GBM cell lines with the same dedifferentiation protocol and compared to 2 patient-derived GSC lines as reference. We gathered existing data on GSC markers to perform an extensive morphological, transcriptional and protein expression characterization. This analysis validated our models while defining their phenotypes and providing support for potential common therapeutic target.
Dedifferentiation protocol led to rapid gliosphere formation in the 4 studied cell lines. Morphological study confirmed that dedifferentiated gliospheres looked like BTIC, with similar size range. Gliosphere areas were highly variable within a given sample, cells being able to form large aggregates, particularly in U-251MG cell line. We hypothesize that the formation of large gliospheres could be associated with higher aggressiveness. Indeed, in the core of huge gliospheres, cells would lack nutrients and oxygen. Hypoxia has been demonstrated to reinforce cancer cell aggressiveness leading to chemotherapy and radiotherapy resistance [32]. Gliosphere size may also be linked to proliferation capacity. More proliferative cells would form larger gliospheres and would be more susceptible to produce aggressive tumors. Given these hypotheses, we believe that morphometric characteristics should be considered for new therapy evaluation.
A first validation of dedifferentiated GS stemness was their ability to self-renew after dissociation. Separating GS constituting cells also reveals their heterogeneity, with GSC-like cells coexisting with more differentiated cells that became adherent when separated from the GS structure. The 4 dedifferentiated GSC models, as well as BTIC, displayed differences in terms of proliferation characteristics. As anticipated during morphological studies, “second generation” T98-G GS were rare and composed of few cells, with a low increase in size over time. A similar profile could be observed for U-251MG, which was contradictory with the large gliospheres formed directly after dedifferentiation. We hypothesize that such spheroids are rather caused by individual GS aggregation than by high proliferation induced by dedifferentiation. Both cell lines may have a quiescent profile, which may be interesting for further pharmacological studies as quiescent cells can resist to conventional therapies [33]. On the contrary, U-118MG and U-87MG reform gliospheres of similar sizes and shapes after dissociation, even with low initial cell quantities, which supports their rapid proliferation characteristics. Such models would be more aggressive and may escape treatment by rapid resistance acquisition or by population renewal. Such heterogeneity in our models is interesting to study how to overcome resistance in GSC and to look for new therapeutic targets that could restore conventional therapy sensitivity.
By screening of 31 genes involved in GSC identification, maintenance or in glioblastoma therapeutic resistance, we confirmed similar characteristics between dedifferentiated cells and GSC and suggested relevant markers for protein studies. Indeed, each model expresses several genes associated with stemness and dedifferentiation increased expression of important stem cell genes, such as SOX2, NANOG, POU5F1 (OCT3/4), ITGA6 and SSEA1. At the protein level, the increase in Nestin, a GSC marker, supports cell stemness in dedifferentiated gliospheres. Flow cytometry pointed out the presence of two other important GSC markers: A2B5 and integrin α-6 (ITGA6). Strikingly, expression of these proteins were enhanced by dedifferentiation which supports GSC characteristics acquisition with this protocol. A2B5 has been associated with GSC survival, self-renewal and proliferation [34] as well as integrin α6 [35], which is also linked to radiotherapy resistance in mesenchymal GSC [36]. Altogether, these markers’ enhancement suggests a resistant GSC profile that remains to be determined.
Mesenchymal GSC are the most challenging therapeutic target as they survive conventional therapies [8,37]. The studied cell lines presented a mesenchymal oriented profile, in line with the literature on BTIC 12m and 25m [29]. Such characteristics were emphasized by dedifferentiation that increased markers associated with EMT or with mesenchymal GSC, such as CD44, integrin α-5 (ITGA5) or FN1. CD44 is particularly important in our models as it was highly expressed and increased after dedifferentiation, both at the transcriptional and protein levels. CD44 is a mesenchymal GSC marker, a receptor for extracellular matrix components, known to be involved in invasiveness and therapeutic resistance [38]. CD44 is a promising target, its inhibition in GSC leading to reduced migration and invasion with an increased survival in mice [39]. It was also proved that CD44 promotes GSC and aggressive growth when associated with its ligand, osteopontin. Our RT-qPCR data showed a moderate to high expression of SPP1 gene (coding for osteopontin) in 4/6 GSC models. Particularly, both CD44 and SPP1 were increased in dedifferentiated U-251MG which may explain their rapid growth. Conversely, proneural phenotype associated markers were weakly expressed. Despite an apparent increase in RT-qPCR, Olig2 was very weakly detected by immunocytochemistry. Similarly, CD133 mRNA expression was low but mainly conserved in dedifferentiated cells and the percentage of positive cells decreased in flow cytometry. It is possible that few dedifferentiated cells expressed CD133, implying heterogeneity within gliospheres. Considering the high CD44 expression, some GSCs expressed both CD133 and CD44, having an intermediate phenotype, between mesenchymal and proneural, which combines both profile complexity. Such phenotype have been associated with large, invasive tumors, presenting rapid growth [11]. CD133 is the most known proneural GSC marker and has been used for a long time as a GSC enrichment marker. Indeed, it appeared essential for GSC maintenance [40] but there is controversy about its use as the sole GSC marker [11]. In our study, weak CD133 expression and GSC diversity support the use of multi-marker and multi-method analysis as the key stone for any GSC study.
Heterogeneity is typical in glioblastoma and particularly in GSC. Due to their plasticity, GSC can easily reshape intracellular pathways to adjust to their environment. Our dedifferentiated and patient derived GSC showed common phenotypic characteristics and stem cell transcriptional patterns, while having their specificities. The 3D GSC organization can be responsible for intra-gliosphere heterogeneity, leading to different cell phenotypes depending on their location. Indeed, hypoxia is known to support GSC phenotype [41] and to increase the expression of several GSC markers such as Sox2 [42] and CD133 [11], but also CD44 under severe hypoxia [39]. Consequently, dedifferentiation process, yet applied to a uniform cell line, may have several intracellular consequences or leads to cell specificities within gliospheres. In vivo, therapies must be effective on high intratumoral cell heterogeneity. If all the cells composing gliospheres may not be GSC, some are definitely stem-like cells. Such diversity in differentiation is mimicking in vivo conditions and gliosphere treatment may induce clonal selection or phenotype drifting. For this reason, future pharmacological studies must include post-treatment analysis of key marker expression such as CD44, CD133, CXCR4, A2B5 and integrin α6. Even if in vitro GSC models lack microenvironment effect, our heterogeneous 3D cultures are suitable to study the effect of inhibitory molecules on complex GSC. Further validation of these GSC models would require in vivo tumorigenicity studies. Tumor growth can be assessed after subcutaneous [14] or intracranial injection [29]. Animal survival and tumor characteristics, such as invasiveness, could be associated with the marker expression profile of GSC models. Moreover, serial transplantation of dedifferentiated GSC would validate their cancer stem cell phenotype, as the number of cells required to induce a tumor can be correlated with the frequency of CSC (Cancer Stem Cells) in a sample [43].
Our gliosphere characterization supports the choice of CXCR4-associated ligand and receptors as therapeutic targets. Indeed, many genes involved in this signaling pathway were more expressed after dedifferentiation. Particularly, MIF and its main receptor, CD74 were highly present at the transcriptional level, but CD74 positive population was rare on flow cytometry. Considering high MIF mRNA expression, its recognition by our GSC may involve secondary receptors such as CXCR4 or CD44. Indeed, CD44, highly expressed in our models, is also able to form complex heteromers for MIF recognition with CXCR4, CXCR7 and CXCR2 [44]. MIF inhibition in GSC decreases stemness, increases radiotherapy sensitivity and induces apoptosis [45]. To confirm MIF as a potential target in our GSC models, future studies must include MIF analysis. Among MIF receptors, CXCR4 appears to have a key role in glioblastoma, which is confirmed in our study. CXCR4 is necessary for GSC phenotype and is associated with EMT, treatment resistance and angiogenesis [25]. Given its importance in tumoral aggressiveness, several inhibitors have been tested on glioblastoma cells. CXCR4 inhibition in GSC decreases self-renewal ability and induces differentiation [46]. Our study provides support to CXCR4 importance in GSC as CXCR4 expression was increased after dedifferentiation in all studied models and was highly expressed in BTIC. Consequently, MIF or CXCR4 inhibitors appear relevant in future GSC targeting strategies.
Conclusion
In summary, we demonstrate that dedifferentiation of 4 commercially available GBM cell lines is an easy way to obtain glioblastoma stem cells. Dedifferentiated cells grow as gliospheres in culture with characteristics closely resembling 2 patient-derived GSC (BTIC) such as self-renewal and stemness marker expression. Analysis of more than 30 GSC and GBM markers revealed a mesenchymal, probably therapy-resistant, GSC phenotype and supports CXCR4 therapeutic targeting in GSC with these 6 studied GSC models. Models have common phenotypic characteristics, but each model has its own specificities in terms of marker expression, gliosphere morphology and growth which may lead to different therapeutic response. To conclude, dedifferentiated cells are robust GSC models and the variability of the 4 models presented in this study would be interesting to evaluate new therapies.
Acknowledgements
The authors acknowledge Dr. H. Artee Luchman and Pr. Samuel Weiss from the Hotchkiss Brain Institute for BTIC supply. We also thank Jérôme Cayon, Lydie Bonneau and Catherine Guillet from PACeM (Plateforme d’Analyse Cellulaire et Moléculaire) as well as Rodolphe Perrot from SCIAM (Service Commun d’Imageries et d’Analyses Microscopiques). Finally, we thank Julien Chaigneau (HIFIH) for his help in image analysis. This study was supported by “Ligue contre le Cancer” (2019 funding), “Université d’Angers” and “CHU d’Angers”, all located in France.
Disclosure of conflict of interest
None.
Abbreviations
- ATCC
American Type Culture Collection
- BTIC
Brain Tumor Initiating Cells
- AC
Adherent cells
- CSC
Cancer Stem Cells
- EMT
Epithelial-Mesenchymal Transition
- FC
Flow Cytometry
- GS
Gliospheres
- GSC
Glioblastoma Stem Cell
- GBM
Glioblastoma
- NSC
Neural Stem Cell
- PN
Proneural
- MES
Mesenchymal
- RT-qPCR
Reverse Transcription-quantitative Polymerase Chain Reaction
Supporting Information
References
- 1.Miller KD, Ostrom QT, Kruchko C, Patil N, Tihan T, Cioffi G, Fuchs HE, Waite KA, Jemal A, Siegel RL, Barnholtz-Sloan JS. Brain and other central nervous system tumor statistics, 2021. CA Cancer J Clin. 2021;71:381–406. doi: 10.3322/caac.21693. [DOI] [PubMed] [Google Scholar]
- 2.Crivii CB, Bosca AB, Melincovici CS, Constantin AM, Mărginean M, Dronca E, Sufletel R, Gonciar D, Bungărdean M, Sovrea A. Glioblastoma microenvironment and cellular interactions. Cancers (Basel) 2022;14:1092. doi: 10.3390/cancers14041092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Biserova K, Jakovlevs A, Uljanovs R, Strumfa I. Cancer stem cells: significance in origin, pathogenesis and treatment of glioblastoma. Cells. 2021;10:621. doi: 10.3390/cells10030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auffinger B, Spencer D, Pytel P, Ahmed AU, Lesniak MS. The role of glioma stem cells in chemotherapy resistance and glioblastoma multiforme recurrence. Expert Rev Neurother. 2015;15:741–752. doi: 10.1586/14737175.2015.1051968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Steponaitis G, Tamasauskas A. Mesenchymal and proneural subtypes of glioblastoma disclose branching based on GSC associated signature. Int J Mol Sci. 2021;22:4964. doi: 10.3390/ijms22094964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Halliday J, Helmy K, Pattwell SS, Pitter KL, LaPlant Q, Ozawa T, Holland EC. In vivo radiation response of proneural glioma characterized by protective p53 transcriptional program and proneural-mesenchymal shift. Proc Natl Acad Sci U S A. 2014;111:5248–5253. doi: 10.1073/pnas.1321014111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bhat KPL, Balasubramaniyan V, Vaillant B, Ezhilarasan R, Hummelink K, Hollingsworth F, Wani K, Heathcock L, James JD, Goodman LD, Conroy S, Long L, Lelic N, Wang S, Gumin J, Raj D, Kodama Y, Raghunathan A, Olar A, Joshi K, Pelloski CE, Heimberger A, Kim SH, Cahill DP, Rao G, Den Dunnen WFA, Boddeke HWGM, Phillips HS, Nakano I, Lang FF, Colman H, Sulman EP, Aldape K. Mesenchymal differentiation mediated by NF-κB promotes radiation resistance in glioblastoma. Cancer Cell. 2013;24:331–346. doi: 10.1016/j.ccr.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mao P, Joshi K, Li J, Kim SH, Li P, Santana-Santos L, Luthra S, Chandran UR, Benos PV, Smith L, Wang M, Hu B, Cheng SY, Sobol RW, Nakano I. Mesenchymal glioma stem cells are maintained by activated glycolytic metabolism involving aldehyde dehydrogenase 1A3. Proc Natl Acad Sci U S A. 2013;110:8644–8649. doi: 10.1073/pnas.1221478110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenkiewicz M, Li N, Singh SK. Culture and isolation of brain tumor initiating cells. Curr Protoc Stem Cell Biol. 2009;11:3.3.1–3.3.10. doi: 10.1002/9780470151808.sc0303s11. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Sun T, Hu J, Zhang R, Rao Y, Wang S, Chen R, McLendon RE, Friedman AH, Keir ST, Bigner DD, Li QJ, Wang H, Wang XF. miR-33a promotes glioma-initiating cell self-renewal via PKA and NOTCH pathways. J Clin Invest. 2014;124:4489–4502. doi: 10.1172/JCI75284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brown DV, Filiz G, Daniel PM, Hollande F, Dworkin S, Amiridis S, Kountouri N, Ng W, Morokoff AP, Mantamadiotis T. Expression of CD133 and CD44 in glioblastoma stem cells correlates with cell proliferation, phenotype stability and intra-tumor heterogeneity. PLoS One. 2017;12:e0172791. doi: 10.1371/journal.pone.0172791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Z, Wang HW, Wang S, Fan L, Feng S, Cai X, Peng C, Wu X, Lu J, Chen D, Chen Y, Wu W, Lu D, Liu N, You Y, Wang H. USP9X deubiquitinates ALDH1A3 and maintains mesenchymal identity in glioblastoma stem cells. J Clin Invest. 2019;129:2043–2055. doi: 10.1172/JCI126414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Seidel S, Garvalov BK, Acker T. Isolation and culture of primary glioblastoma cells from human tumor specimens. In: Rich IN, editor. Stem cell protocols. New York: Springer New York; 2015. pp. 263–275. [DOI] [PubMed] [Google Scholar]
- 14.Ishii H, Mimura Y, Zahra MH, Katayama S, Hassan G, Afify SM, Seno M. Isolation and characterization of cancer stem cells derived from human glioblastoma. Am J Cancer Res. 2021;11:441–457. [PMC free article] [PubMed] [Google Scholar]
- 15.Yasmin IA, Mohana Sundaram S, Banerjee A, Varier L, Dharmarajan A, Warrier S. Netrin-like domain of sFRP4, a Wnt antagonist inhibits stemness, metastatic and invasive properties by specifically blocking MMP-2 in cancer stem cells from human glioma cell line U87MG. Exp Cell Res. 2021;409:112912. doi: 10.1016/j.yexcr.2021.112912. [DOI] [PubMed] [Google Scholar]
- 16.Peixoto J, Janaki-Raman S, Schlicker L, Schmitz W, Walz S, Winkelkotte AM, Herold-Mende C, Soares P, Schulze A, Lima J. Integrated metabolomics and transcriptomics analysis of monolayer and neurospheres from established glioblastoma cell lines. Cancers (Basel) 2021;13:1327. doi: 10.3390/cancers13061327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Prager BC, Wu Q, Kim LJY, Gimple RC, Shi Y, Yang K, Morton AR, Zhou W, Zhu Z, Obara EAA, Miller TE, Song A, Lai S, Hubert CG, Jin X, Huang Z, Fang X, Dixit D, Tao W, Zhai K, Chen C, Dong Z, Zhang G, Dombrowski SM, Hamerlik P, Mack SC, Bao S, Rich JN. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell. 2018;22:514–528. e5. doi: 10.1016/j.stem.2018.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erhart F, Blauensteiner B, Zirkovits G, Printz D, Soukup K, Klingenbrunner S, Fischhuber K, Reitermaier R, Halfmann A, Lötsch D, Spiegl-Kreinecker S, Berger W, Visus C, Dohnal A. Gliomasphere marker combinatorics: multidimensional flow cytometry detects CD44+/CD133+/ITGA6+/CD36+ signature. J Cell Mol Med. 2019;23:281–292. doi: 10.1111/jcmm.13927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Flüh C, Chitadze G, Adamski V, Hattermann K, Synowitz M, Kabelitz D, Held-Feindt J. NKG2D ligands in glioma stem-like cells: expression in situ and in vitro. Histochem Cell Biol. 2018;149:219–233. doi: 10.1007/s00418-018-1633-5. [DOI] [PubMed] [Google Scholar]
- 20.Lee JW, Sung JS, Park YS, Chung S, Kim YH. Isolation of spheroid-forming single cells from gastric cancer cell lines: enrichment of cancer stem-like cells. Biotechniques. 2018;65:197–203. doi: 10.2144/btn-2018-0046. [DOI] [PubMed] [Google Scholar]
- 21.Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 23.Gu Z. Complex heatmap visualization. IMeta. 2022;1:e43. doi: 10.1002/imt2.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 25.Richardson PJ. CXCR4 and glioblastoma. Anticancer Agents Med Chem. 2015;16:59–74. doi: 10.2174/1871520615666150824153032. [DOI] [PubMed] [Google Scholar]
- 26.He J, Liu Y, Zhu T, Zhu J, DiMeco F, Vescovi AL, Heth JA, Muraszko KM, Fan X, Lubman DM. CD90 is identified as a candidate marker for cancer stem cells in primary high-grade gliomas using tissue microarrays. Mol Cell Proteomics. 2012;11:M111.010744. doi: 10.1074/mcp.M111.010744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou Y, Meng X, He W, Li X, Zhao R, Dong C, Yuan D, Yang J, Zhang R, Shi G, Huang Y, Liu J, Liu J, Liu S, Fu P, Sun M. USF1/CD90 signaling in maintaining glioblastoma stem cells and tumor-associated macrophages adhesion. Neuro Oncol. 2022;24:1482–1493. doi: 10.1093/neuonc/noac063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woo SR, Oh YT, An JY, Kang BG, Nam DH, Joo KM. Glioblastoma specific antigens, GD2 and CD90, are not involved in cancer stemness. Anat Cell Biol. 2015;48:44–53. doi: 10.5115/acb.2015.48.1.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cusulin C, Chesnelong C, Bose P, Bilenky M, Kopciuk K, Chan JA, Cairncross JG, Jones SJ, Marra MA, Luchman HA, Weiss S. Precursor states of brain tumor initiating cell lines are predictive of survival in xenografts and associated with glioblastoma subtypes. Stem Cell Rep. 2015;5:1–9. doi: 10.1016/j.stemcr.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kelly JJ, Stechishin O, Chojnacki A, Lun X, Sun B, Senger DL, Forsyth P, Auer RN, Dunn JF, Cairncross JG, Parney IF, Weiss S. Proliferation of human glioblastoma stem cells occurs independently of exogenous mitogens. Stem Cells. 2009;27:1722–1733. doi: 10.1002/stem.98. [DOI] [PubMed] [Google Scholar]
- 31.van Bodegraven EJ, van Asperen JV, Robe PAJ, Hol EM. Importance of GFAP isoform-specific analyses in astrocytoma. Glia. 2019;67:1417–1433. doi: 10.1002/glia.23594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cosse JP, Ronvaux M, Ninane N, Raes MJ, Michiels C. Hypoxia-induced decrease in p53 protein level and increase in c-jun DNA binding activity results in cancer cell resistance to etoposide. Neoplasia. 2009;11:976–986. doi: 10.1593/neo.09632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tejero R, Huang Y, Katsyv I, Kluge M, Lin JY, Tome-Garcia J, Daviaud N, Wang Y, Zhang B, Tsankova NM, Friedel CC, Zou H, Friedel RH. Gene signatures of quiescent glioblastoma cells reveal mesenchymal shift and interactions with niche microenvironment. EBioMedicine. 2019;42:252–269. doi: 10.1016/j.ebiom.2019.03.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Baeza-Kallee N, Bergès R, Soubéran A, Colin C, Denicolaï E, Appay R, Tchoghandjian A, Figarella-Branger D. Glycolipids recognized by A2B5 antibody promote proliferation, migration, and clonogenicity in glioblastoma cells. Cancers (Basel) 2019;11:1267. doi: 10.3390/cancers11091267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, MacSwords J, Wu Q, Vasanji A, McLendon RE, Hjelmeland AB, Rich JN. Integrin Alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010;6:421–432. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stanzani E, Pedrosa L, Bourmeau G, Anezo O, Noguera-Castells A, Esteve-Codina A, Passoni L, Matteoli M, de la Iglesia N, Seano G, Martínez-Soler F, Tortosa A. Dual role of integrin Alpha-6 in glioblastoma: supporting stemness in proneural stem-like cells while inducing radioresistance in mesenchymal stem-like cells. Cancers (Basel) 2021;13:3055. doi: 10.3390/cancers13123055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minata M, Audia A, Shi J, Lu S, Bernstock J, Pavlyukov MS, Das A, Kim SH, Shin YJ, Lee Y, Koo H, Snigdha K, Waghmare I, Guo X, Mohyeldin A, Gallego-Perez D, Wang J, Chen D, Cheng P, Mukheef F, Contreras M, Reyes JF, Vaillant B, Sulman EP, Cheng SY, Markert JM, Tannous BA, Lu X, Kango-Singh M, Lee LJ, Nam DH, Nakano I, Bhat KP. Phenotypic plasticity of invasive edge glioma stem-like cells in response to ionizing radiation. Cell Rep. 2019;26:1893–1905. e7. doi: 10.1016/j.celrep.2019.01.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wolf KJ, Shukla P, Springer K, Lee S, Coombes JD, Choy CJ, Kenny SJ, Xu K, Kumar S. A mode of cell adhesion and migration facilitated by CD44-dependent microtentacles. Proc Natl Acad Sci U S A. 2020;117:11432–11443. doi: 10.1073/pnas.1914294117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nishikawa M, Inoue A, Ohnishi T, Yano H, Ozaki S, Kanemura Y, Suehiro S, Ohtsuka Y, Kohno S, Ohue S, Shigekawa S, Watanabe H, Kitazawa R, Tanaka J, Kunieda T. Hypoxia-induced phenotypic transition from highly invasive to less invasive tumors in glioma stem-like cells: significance of CD44 and osteopontin as therapeutic targets in glioblastoma. Transl Oncol. 2021;14:101137. doi: 10.1016/j.tranon.2021.101137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brescia P, Ortensi B, Fornasari L, Levi D, Broggi G, Pelicci G. CD133 is essential for glioblastoma stem cell maintenance. Stem Cells. 2013;31:857–869. doi: 10.1002/stem.1317. [DOI] [PubMed] [Google Scholar]
- 41.Karsy M, Guan J, Jensen R, Huang LE, Colman H. The impact of hypoxia and mesenchymal transition on glioblastoma pathogenesis and cancer stem cells regulation. World Neurosurg. 2016;88:222–236. doi: 10.1016/j.wneu.2015.12.032. [DOI] [PubMed] [Google Scholar]
- 42.Bhagat M, Palanichamy JK, Ramalingam P, Mudassir M, Irshad K, Chosdol K, Sarkar C, Seth P, Goswami S, Sinha S, Chattopadhyay P. HIF-2α mediates a marked increase in migration and stemness characteristics in a subset of glioma cells under hypoxia by activating an Oct-4/Sox-2-Mena (INV) axis. Int J Biochem Cell Biol. 2016;74:60–71. doi: 10.1016/j.biocel.2016.02.017. [DOI] [PubMed] [Google Scholar]
- 43.Rycaj K, Tang DG. Cell-of-origin of cancer versus cancer stem cells: assays and interpretations. Cancer Res. 2015;75:4003–4011. doi: 10.1158/0008-5472.CAN-15-0798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jankauskas SS, Wong DWL, Bucala R, Djudjaj S, Boor P. Evolving complexity of MIF signaling. Cell Signal. 2019;57:76–88. doi: 10.1016/j.cellsig.2019.01.006. [DOI] [PubMed] [Google Scholar]
- 45.Lee SH, Kwon HJ, Park S, Kim CI, Ryu H, Kim SS, Park JB, Kwon JT. Macrophage migration inhibitory factor (MIF) inhibitor 4-IPP downregulates stemness phenotype and mesenchymal trans-differentiation after irradiation in glioblastoma multiforme. PLoS One. 2021;16:e0257375. doi: 10.1371/journal.pone.0257375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gravina GL, Mancini A, Colapietro A, Vitale F, Vetuschi A, Pompili S, Rossi G, Marampon F, Richardson PJ, Patient L, Patient L, Burbidge S, Festuccia C. The novel CXCR4 antagonist, PRX177561, reduces tumor cell proliferation and accelerates cancer stem cell differentiation in glioblastoma preclinical models. Tumour Biol. 2017;39:1010428317695528. doi: 10.1177/1010428317695528. [DOI] [PubMed] [Google Scholar]
- 47.Tchoghandjian A, Baeza N, Colin C, Cayre M, Metellus P, Beclin C, Ouafi k L, Figarella-Branger D. A2B5 cells from human glioblastoma have cancer stem cell properties. Brain Pathol. 2010;20:211–21. doi: 10.1111/j.1750-3639.2009.00269.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sun T, Chen G, Li Y, Xie X, Zhou Y, Du Z. Aggressive invasion is observed in CD133-/A2B5+ glioma-initiating cells. Oncol Lett. 2015;10:3399–3406. doi: 10.3892/ol.2015.3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jin X, Kim LJY, Wu Q, Wallace LC, Prager BC, Sanvoranart T, Gimple RC, Wang X, Mack SC, Miller TE, Huang P, Valentim CL, Zhou QG, Barnholtz-Sloan JS, Bao S, Sloan AE, Rich JN. Targeting glioma stem cells through combined BMI1 and EZH2 inhibition. Nat Med. 2017;23:1352–1361. doi: 10.1038/nm.4415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Flamier A, Abdouh M, Hamam R, Barabino A, Patel N, Gao A, Hanna R, Bernier G. Off-target effect of the BMI1 inhibitor PTC596 drives epithelial-mesenchymal transition in glioblastoma multiforme. NPJ Precis Oncol. 2020;4:1. doi: 10.1038/s41698-019-0106-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brown DV, Daniel PM, D’Abaco GM, Gogos A, Ng W, Morokoff AP, Mantamadiotis T. Coexpression analysis of CD133 and CD44 identifies proneural and mesenchymal subtypes of glioblastoma multiforme. Oncotarget. 2015;6:6267–6280. doi: 10.18632/oncotarget.3365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu G, Yuan X, Zeng Z, Tunici P, Ng H, Abdulkadir IR, Lu L, Irvin D, Black KL, Yu JS. Analysis of gene expression and chemoresistance of CD133+ cancer stem cells in glioblastoma. Mol Cancer. 2006;5:67. doi: 10.1186/1476-4598-5-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- 54.Hale JS, Otvos B, Sinyuk M, Alvarado AG, Hitomi M, Stoltz K, Wu Q, Flavahan W, Levison B, Johansen ML, Schmitt D, Neltner JM, Huang P, Ren B, Sloan AE, Silverstein RL, Gladson CL, DiDonato JA, Brown JM, McIntyre T, Hazen SL, Horbinski C, Rich JN, Lathia JD. Cancer stem cell-specific scavenger receptor CD36 drives glioblastoma progression. Stem Cells. 2014;32:1746–1758. doi: 10.1002/stem.1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Presti M, Mazzon E, Basile M, Petralia M, Bramanti A, Colletti G, Bramanti P, Nicoletti F, Fagone P. Overexpression of macrophage migration inhibitory factor and functionally-related genes, D-DT, CD74, CD44, CXCR2 and CXCR4, in glioblastoma. Oncol Lett. 2018;16:2881–2886. doi: 10.3892/ol.2018.8990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gött H, Nagl J, Hagedorn F, Thomas S, Schwarm F, Uhl E, Kolodziej M. ZEB1 induces N-cadherin expression in human glioblastoma and may alter patient survival. Mol Clin Oncol. 2022;17:123. doi: 10.3892/mco.2022.2556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chen Q, Cai J, Jiang C. CDH2 expression is of prognostic significance in glioma and predicts the efficacy of temozolomide therapy in patients with glioblasotma. Oncol Lett. 2018;15:7415–7422. doi: 10.3892/ol.2018.8227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gatti M, Pattarozzi A, Bajetto A, Würth R, Daga A, Fiaschi P, Zona G, Florio T, Barbieri F. Inhibition of CXCL12/CXCR4 autocrine/paracrine loop reduces viability of human glioblastoma stem-like cells affecting self-renewal activity. Toxicology. 2013;314:209–220. doi: 10.1016/j.tox.2013.10.003. [DOI] [PubMed] [Google Scholar]
- 59.Infanger DW, Cho Y, Lopez BS, Mohanan S, Liu SC, Gursel D, Boockvar JA, Fischbach C. Glioblastoma stem cells are regulated by interleukin-8 signaling in a tumoral perivascular niche. Cancer Res. 2013;73:7079–7089. doi: 10.1158/0008-5472.CAN-13-1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng X, Xie Q, Li S, Zhang W. CXCR4-positive subset of glioma is enriched for cancer stem cells. Oncol Res. 2011;19:555–561. doi: 10.3727/096504012x13340632812631. [DOI] [PubMed] [Google Scholar]
- 61.Ehtesham M, Mapara KY, Stevenson CB, Thompson RC. CXCR4 mediates the proliferation of glioblastoma progenitor cells. Cancer Lett. 2009;274:305–312. doi: 10.1016/j.canlet.2008.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Walters MJ, Ebsworth K, Berahovich RD, Penfold ME, Liu SC, Al Omran R, Kioi M, Chernikova SB, Tseng D, Mulkearns-Hubert EE, Sinyuk M, Ransohoff RM, Lathia JD, Karamchandani J, Kohrt HE, Zhang P, Powers JP, Jaen JC, Schall TJ, Merchant M, Recht L, Brown JM. Inhibition of CXCR7 extends survival following irradiation of brain tumours in mice and rats. Br J Cancer. 2014;110:1179–1188. doi: 10.1038/bjc.2013.830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hattermann K, Held-Feindt J, Lucius R, Müerköster SS, Penfold ME, Schall TJ, Mentlein R. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res. 2010;70:3299–3308. doi: 10.1158/0008-5472.CAN-09-3642. [DOI] [PubMed] [Google Scholar]
- 64.Hattermann K, Mentlein R. An infernal trio: the chemokine CXCL12 and its receptors CXCR4 and CXCR7 in tumor biology. Ann Anat. 2013;195:103–110. doi: 10.1016/j.aanat.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 65.Takashima Y, Kawaguchi A, Yamanaka R. Promising prognosis marker candidates on the status of epithelial-mesenchymal transition and glioma stem cells in glioblastoma. Cells. 2019;8:1312. doi: 10.3390/cells8111312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Seifert C, Balz E, Herzog S, Korolev A, Gaßmann S, Paland H, Fink MA, Grube M, Marx S, Jedlitschky G, Tzvetkov MV, Rauch BH, Schroeder HWS, Bien-Möller S. PIM1 inhibition affects glioblastoma stem cell behavior and kills glioblastoma stem-like cells. Int J Mol Sci. 2021;22:11126. doi: 10.3390/ijms222011126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Shevchenko V, Arnotskaya N, Pak O, Sharma A, Sharma HS, Khotimchenko Y, Bryukhovetskiy A, Bryukhovetskiy I. Molecular determinants of the interaction between glioblastoma CD133+ cancer stem cells and the extracellular matrix. Int Rev Neurobiol. 2020;151:155–169. doi: 10.1016/bs.irn.2020.03.005. [DOI] [PubMed] [Google Scholar]
- 68.Bao S, Wu Q, Li Z, Sathornsumetee S, Wang H, McLendon RE, Hjelmeland AB, Rich JN. Targeting cancer stem cells through L1CAM suppresses glioma growth. Cancer Res. 2008;68:6043–6048. doi: 10.1158/0008-5472.CAN-08-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fukaya R, Ohta S, Yaguchi T, Matsuzaki Y, Sugihara E, Okano H, Saya H, Kawakami Y, Kawase T, Yoshida K, Toda M. MIF maintains the tumorigenic capacity of brain tumor-initiating cells by directly inhibiting p53. Cancer Res. 2016;76:2813–2823. doi: 10.1158/0008-5472.CAN-15-1011. [DOI] [PubMed] [Google Scholar]
- 70.Niu CS, Li DX, Liu YH, Fu XM, Tang SF, Li J. Expression of NANOG in human gliomas and its relationship with undifferentiated glioma cells. Oncol Rep. 2011;26:593–601. doi: 10.3892/or.2011.1308. [DOI] [PubMed] [Google Scholar]
- 71.Morfouace M, Lalier L, Oliver L, Cheray M, Pecqueur C, Cartron PF, Vallette FM. Control of glioma cell death and differentiation by PKM2-Oct4 interaction. Cell Death Dis. 2014;5:e1036. doi: 10.1038/cddis.2013.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ikushima H, Todo T, Ino Y, Takahashi M, Saito N, Miyazawa K, Miyazono K. Glioma-initiating cells retain their tumorigenicity through integration of the Sox axis and Oct4 protein. J Biol Chem. 2011;286:41434–41441. doi: 10.1074/jbc.M111.300863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Bouchart C, Trépant A, Hein M, Van Gestel D, Demetter P. Prognostic impact of glioblastoma stem cell markers OLIG2 and CCND2. Cancer Med. 2020;9:1069–1078. doi: 10.1002/cam4.2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chow KH, Park HJ, George J, Yamamoto K, Gallup AD, Graber JH, Chen Y, Jiang W, Steindler DA, Neilson EG, Kim BYS, Yun K. S100A4 is a biomarker and regulator of glioma stem cells that is critical for mesenchymal transition in glioblastoma. Cancer Res. 2017;77:5360–5373. doi: 10.1158/0008-5472.CAN-17-1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chesnelong C, Hao X, Cseh O, Wang AY, Luchman HA, Weiss S. SLUG directs the precursor state of human brain tumor stem cells. Cancers (Basel) 2019;11:1635. doi: 10.3390/cancers11111635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hägerstrand D, He X, Bradic Lindh M, Hoefs S, Hesselager G, Östman A, Nistér M. Identification of a SOX2-dependent subset of tumor- and sphere-forming glioblastoma cells with a distinct tyrosine kinase inhibitor sensitivity profile. Neuro Oncol. 2011;13:1178–1191. doi: 10.1093/neuonc/nor113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pietras A, Katz AM, Ekström EJ, Wee B, Halliday JJ, Pitter KL, Werbeck JL, Amankulor NM, Huse JT, Holland EC. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14:357–369. doi: 10.1016/j.stem.2014.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Son MJ, Woolard K, Nam DH, Lee J, Fine HA. SSEA-1 is an enrichment marker for tumor-initiating cells in human glioblastoma. Cell Stem Cell. 2009;4:440–452. doi: 10.1016/j.stem.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hong X, Jiang F, Kalkanis SN, Zhang ZG, Zhang XP, deCarvalho AC, Katakowski M, Bobbitt K, Mikkelsen T, Chopp M. SDF-1 and CXCR4 are up-regulated by VEGF and contribute to glioma cell invasion. Cancer Lett. 2006;236:39–45. doi: 10.1016/j.canlet.2005.05.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.