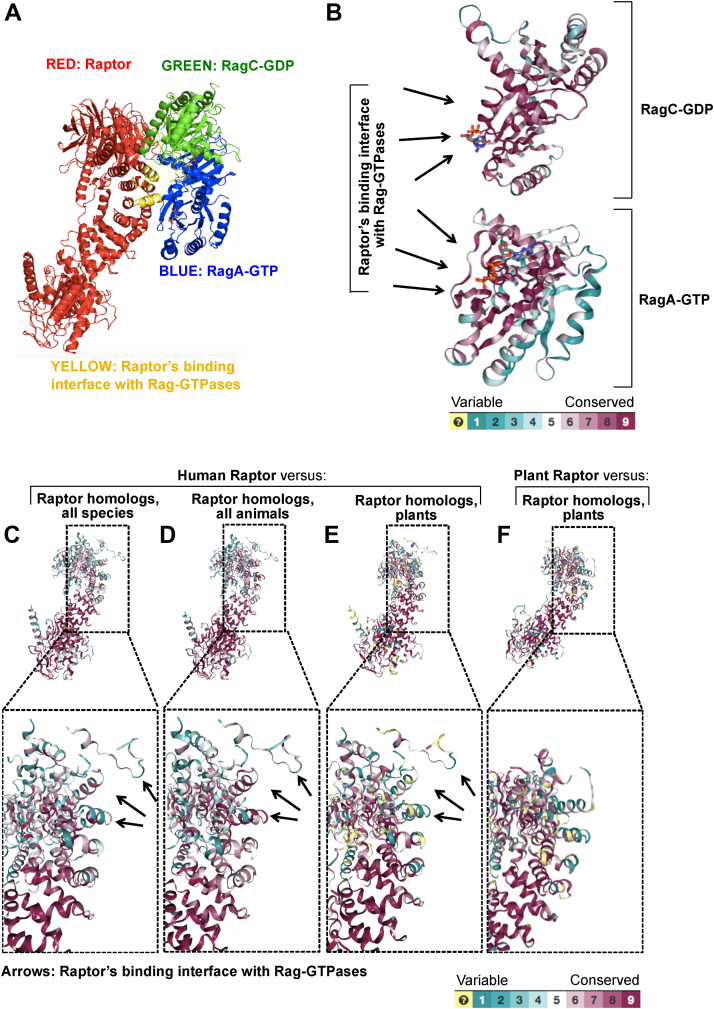
Figure 2.
Conserved interacting regions of Raptor and Rag GTPases highlight the utility of the affinity assay in all mTOR homolog-expressing species except plants.A, the crystal structure of human Raptor with the dimer of human RagA-GTP with RagC-GDP at 3.18 Ångström with the interacting regions between the supercomplex highlighted in yellow. The structure was obtained from PDB file 6U62 (7). A conserved protein sequence analysis comparing human versus all homolog-containing species for RagA and RagC (B) or Raptor (C–E) is superimposed onto PDB 6U62 using ConSurf software. Conserved protein sequences for human Raptor versus Raptor homologs from all species (C), all animals (D), and plants (E) are shown. F, Raptor-conserved protein sequences for Arabidopsis thaliana (plant) versus homologs from all other plants superimposed onto Arabidopsis thaliana Raptor PDB 5WBI (14). Computed conservation scores are assigned color grades 1 to 9, going from variable to conserved, as indicated. Unreliable regions are colored in light yellow. For more info, see Experimental procedures and references therein. mTOR, mechanistic target of rapamycin.