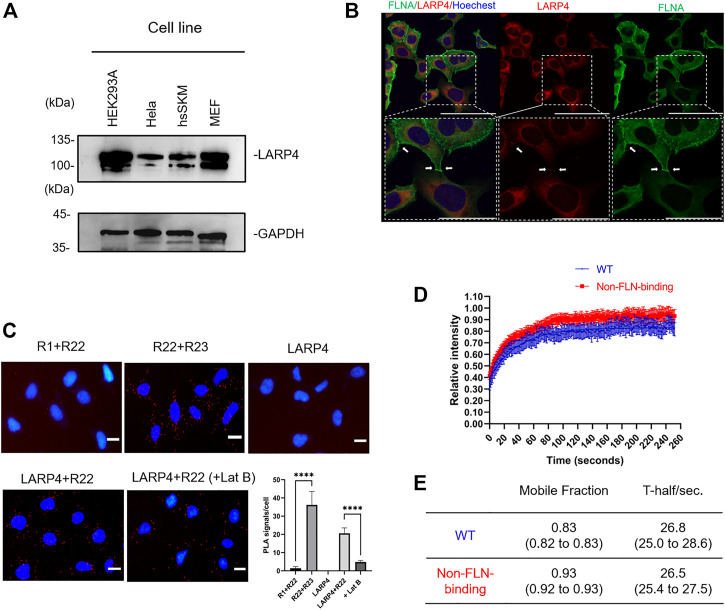
FIGURE 6.
Interaction of LARP4 with FLNA in living cells. (A) Approximately 1.2 × 106 HEK293A, Hela, hsSKM, and MEF cells cells were analyzed by WB using anti-LARP4 antibodies. The lower band is the indicative of the degradation of LARP4 protein. GAPDH is used as a loading control. (B) Co-localization of LARP4 and FLNA in HEK293A (Bar: 100 μm). Enlarged images are shown in the dotted box in white. Bar: 50 μm. White arrows indicate the cell-cell junction. (C) Interactions of LARP4 with FLNA R22 in HEK293A cells visualized by proximity ligation assay (PLA). Representative PLA images where the PLA signal (red) represents close proximity (<40 nm) between two proteins. PLA signal is significantly decreased when cells are treated with 5 μM Latrunculin B for 1 h. Graph shows the quantification of PLA between FLNA R1 and R22, R22 and R23, LARP4, LARP4 and R22 and under Lat B treatment respectively (n = 5). The nucleus was stained by Hoechst (blue). Scale bars are 20 μm ****p < 0.0001 was determined by the two-tailed unpaired Student’s test. (D) FRAP analysis was performed in HEK293A cells transfected GFP-LARP4 WT or GFP-LARP4 F277A. Quantitative analyses of FRAP assay results. Curves depict mean values (±SD) from measurements of at least 6 representative cells (n ≥ 10). WT (blue), non-FLN-binding F277A (red). (E) Fmax, which represents the mobile fraction of the molecule in the bleached region, and τ½, which is the time to recover half of the maximum fluorescence and is inversely correlated to the diffusion coefficient were analyzed by one-phase association model plugged into the Prism software. (95% confidence intervals).