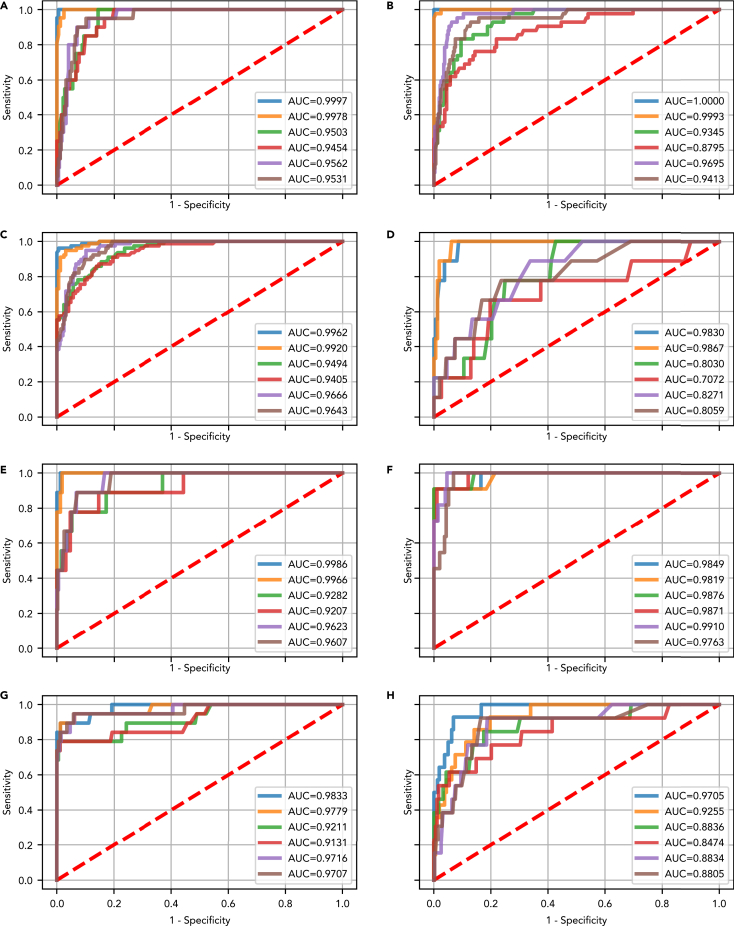
Figure 3.
Test set receiver operating characteristic (ROC) curve and area under the curve (AUC) showing the performance of the best optimized classifier model (selected based on validation results) for detecting a target cancer from the Cohen et al. dataset
The test set results represent a generalizable estimate of performance for DEcancer’s all cancer pipeline. DEcancerP uses proteins-only and DEcancerPDE includes all 39 proteins, DNA-based and epidemiology factors. (Blue) The DEcancerPDE approach for target cancer versus cancer-free test set ROC curve showing performance of optimal model. (Orange) The DEcancerP approach for the target cancer versus cancer-free test set ROC curve. (Green) The DEcancerPDE approach for the target cancer versus other cancers test set ROC curve showing performance of optimal model. (Red) The DEcancerP approach for the target cancer versus other cancers test set ROC curve. (Purple) The DEcancerPDE approach for the target cancer versus other cancers or cancer-free test set ROC curve showing performance of optimal model. (Brown) The DEcancerP approach for the target cancer versus other cancers or cancer-free test set ROC curve.
(A) Target cancer: lung. (Blue) An AUC of 1.00 is achieved with the DEcancerPDE model. (Orange) An AUC of 1.00 is achieved with a 12-protein DEcancerP model. (Green) An AUC of 0.95 is achieved with the DEcancerPDE model. (Red) An AUC of 0.95 is achieved with a 39-protein DEcancerP model. (Purple) An AUC of 0.96 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.95 is achieved with a 22-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 95.24 and 99.39%, respectively.
(B) Target cancer: breast. (Blue) An AUC of 1.00 is achieved with the DEcancerPDE model. (Orange) An AUC of 1.00 is achieved with a 27-protein DEcancerP model. (Green) An AUC of 0.93 is achieved with a DEcancerPDE model. (Red) An AUC of 0.88 is achieved with a 29-protein DEcancerP model. (Purple) An AUC of 0.97 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.94 is achieved with a 26-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 97.62 and 100.00%, respectively.
(C) Target cancer: colorectal. (Blue) An AUC of 1.00 is achieved with the DEcancerPDE model. (Orange) An AUC of 0.99 is achieved with a 22-protein DEcancerP model. (Green) An AUC of 0.95 is achieved with the DEcancerPDE model. (Red) An AUC of 0.94 is achieved with a 22-protein DEcancerP model. (Purple) An AUC of 0.97 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.96 is achieved with a 35-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 94.87 and 99.38%, respectively.
(D) Target cancer: esophageal. (Blue) An AUC of 0.98 is achieved with the DEcancerPDE model. (Orange) An AUC of 0.99 is achieved with an 8-protein DEcancerP model. (Green) An AUC of 0.80 is achieved with the DEcancerPDE. (Red) An AUC of 0.71 achieved with a 23-protein DEcancerP model. (Purple) An AUC of 0.83 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.81 is achieved with a 15-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 88.89 and 94.48%, respectively.
(E) Target cancer: liver. (Blue) An AUC of 1.00 is achieved with the DEcancerPDE model. (Orange) An AUC of 1.00 is achieved with a 23-protein DEcancerP model. (Green) An AUC of 0.93 is achieved with the DEcancerPDE model. (Red) An AUC of 0.92 is achieved with an 8-protein DEcancerP model. (Purple) An AUC of 0.96 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.96 is achieved with a 19-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 88.89 and 98.77%, respectively.
(F) Target cancer: ovarian. (Blue) An AUC of 0.98 is achieved with the DEcancerPDE model. (Orange) An AUC of 0.98 is achieved with a 15-protein DEcancerP model. (Green) An AUC of 0.99 is achieved with the DEcancerPDE model. (Red) An AUC of 0.99 is achieved with a 13-protein DEcancerP model. (Purple) An AUC of 0.99 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.98 is achieved with a 12-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 90.91 and 95.47%, respectively.
(G) Target cancer: pancreatic. (Blue) An AUC of 0.98 is achieved with the DEcancerPDE model. (Orange) An AUC of 0.98 is achieved with a 8-protein DEcancerP model. (Green) An AUC of 0.92 is achieved with the DEcancerPDE model. (Red) An AUC of 0.91 is achieved with a 14-protein DEcancerP model. (Purple) An AUC of 0.97 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.97 is achieved with a 9-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 84.21 and 98.77%, respectively.
(H) Target cancer: gastric. (Blue) An AUC of 0.97 is achieved with the DEcancerPDE model. (Orange) An AUC of 0.93 is achieved with a 17-protein DEcancerP model. (Green) An AUC of 0.88 is achieved with the DEcancerPDE model. (Red) An AUC of 0.85 is achieved with a 19-protein DEcancerP model. (Purple) An AUC of 0.88 is achieved with the DEcancerPDE model. (Brown) An AUC of 0.88 is achieved with a 25-protein DEcancerP model. The optimal sensitivity and specificity corresponding to the top left corner of ROC curve is 85.71 and 93.21%, respectively.