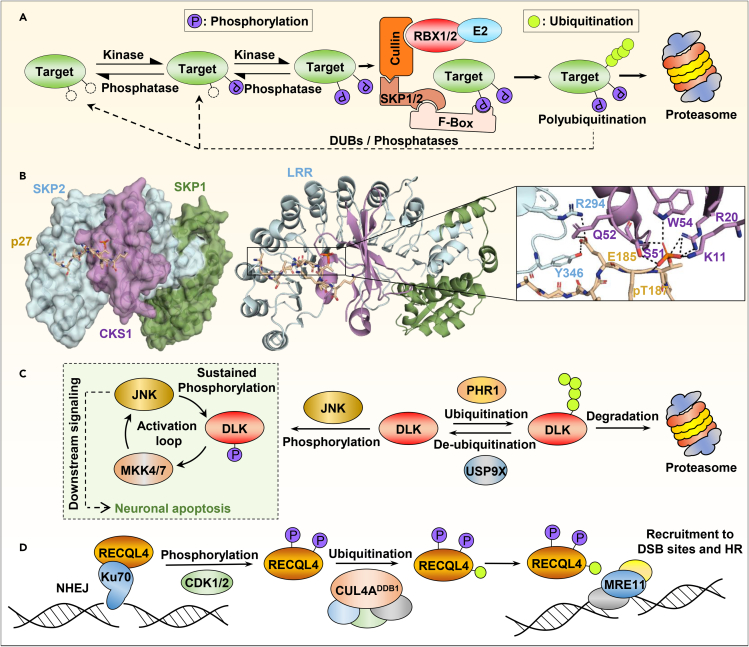
Figure 4.
Model of the interplay between protein phosphorylation and ubiquitination
(A) Phosphorylation of a target protein creates a phosphodegron, recognized by E3 ligase complexes (in this case the SCF or Skip-Cullin-F-box complex family). The F box factor positions the targeted protein in the vicinity of the Cullin ligase (RBX1/2) and the E2 enzyme, leading to its ubiquitination and proteasomal degradation.
(B) Crystal structure of the quaternary complex: SKP1-SKP2-CKS1-Phospho p27Kip1 (PDB: 2AST). Left panel, overall representation of the complex showing the specific positioning of the p27 peptide within CKS1 and SKP2 binding pockets. Right panel, close up view of the phosphorylated p27Kip1 interaction with CKS1 and SKP2. Phosphorylated p27Kip1 intercalates into a CKS1/SKP2 pocket formed by SKP2 leucine-rich repeat (LRR) and CKS1 phospho binding site. pT187 is recognized by CKS1 phospho binding site residues whereas E185 binds to both CKS1 and SKP2. The hydrogen bounds between amino acids are shown at the bottom by the dashed lines.
(C) DLK stability is regulated through the action of the PHR1 E3 ligase and the DUB USP9X. DLK phosphorylation by JNK blocks its ubiquitination and degradation to reinforce the JNK signaling pathway and promote neuronal apoptosis.
(D) Phosphorylation-mediated monoubiquitination of RECQL4 regulates pathway choice of DSB repair. Following DNA damage, RECQL4 associates with the DSB binding protein Ku70 to allow non-homologous end-joining (NHEJ) repair. CDK1/2 phosphorylates RECQL4 which leads to its ubiquitination by CUL4ADDB1 and induces its interaction with MRE11 to promote homologous recombination (HR).