Figure 5.
Crosstalk between methylation or acetylation and ubiquitination
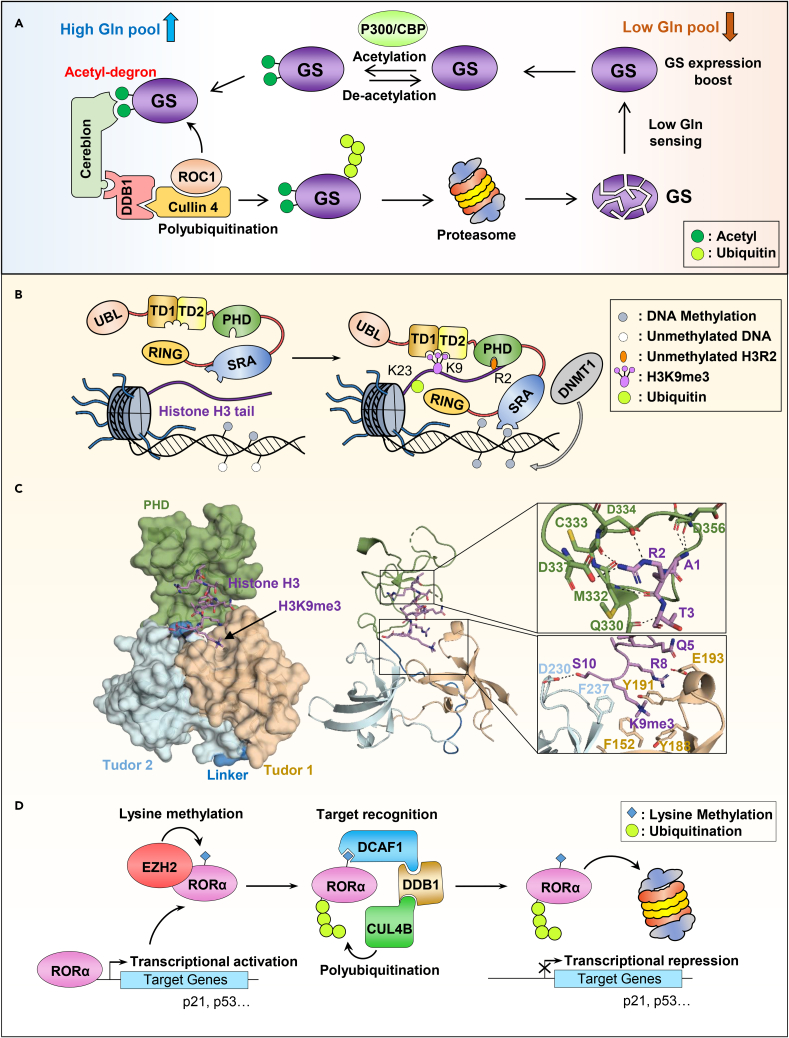
(A) High level of cellular glutamine (Gln) leads to the acetylation of Glutamine synthetase (GS) by P300/CBP. This event creates an acetyl-degron recognized by the CRL4CRBN E3 complex leading to GS polyubiquitination and subsequent degradation by the proteasome.
(B) Bivalent recognition of unmethylated H3R2 and methylated H3K9 by the multidomain E3 ligase, UHRF1, leading to the ubiquitination of H3K23.
(C) Surface representation of the crystal structure of the E3 ubiquitin ligase UHRF1 in complex with histone H3 peptide (PDB: 3ASK). The Tudor domains and the PHD domain (TTD-PHD) used for the crystallization are shown in the left panel. Right panel, zoom in view of the interaction between histone H3 peptide with the PHD domain (top) and the Tudor1/2 domains (bottom). The H3 peptide is composed of two cassettes: cassette 1 encompassing H3R2, is positioned within the PHD acidic pocket; and cassette 2 containing H3K9me3 is recognized by an “aromatic cage” surface within Tudor 1. The hydrogen bounds between amino acids are shown by the dashed lines. The structure shows that the unmodified H3R2 intercalates into an acidic pocket within the PHD finger domain. The Tudor domain 1 accommodated the H3 peptide C-terminus residues into an “aromatic cage” involving H3K9me3 and S10.
(D) Regulation of the RORα nuclear receptor by a methylation/ubiquitination crosstalk. RORα is subjected to monomethylation by the PRC2 methyl-transferase EZH2. This monomethylated RORα is bound by the chromo-domain of DCAF1 recruiting it to the DCAF1/DDB1/CUL4B ubiquitin ligase complex, resulting in the proteasomal degradation of RORα and repression of its target genes.