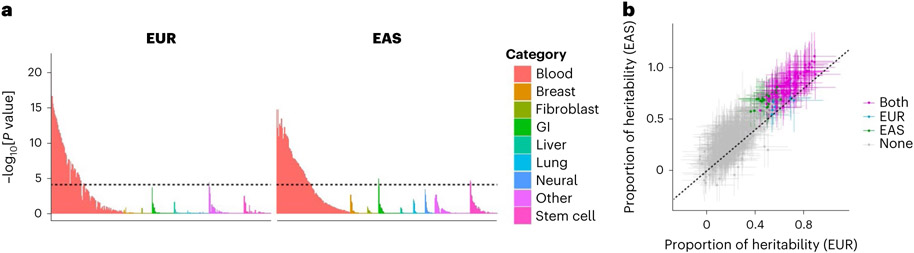
Fig. 5 ∣. S-LDSC analysis suggested similar causal variant distributions in EUR-GWAS and EAS-GWAS.
a, EUR-GWAS and EAS-GWAS results were analyzed by S-LDSC using 707 IMPACT annotations. P values were calculated by block-jackknife implemented in the LDSC software and indicate the significance of non-negative tau (per variant heritability) of each annotation (one-sided test). The color of each annotation indicates its cell-type category. The horizontal dashed line indicates the Bonferroni-corrected P value threshold (0.05/707 = 7.1 × 10−5). b, The estimate and its 95% confidence interval of the heritability proportion explained by the top 5% of IMPACT annotations. Confidence intervals and the P values indicating non-negative tau were estimated via block-jackknife implemented in the LDSC software (one-sided test). The color of each annotation indicates the type of GWAS when a heritability enrichment was significant (P < 0.05/707 = 7.1 × 10−5). ‘Both’ indicates that the annotation was significant in both EUR-GWAS and EAS-GWAS.