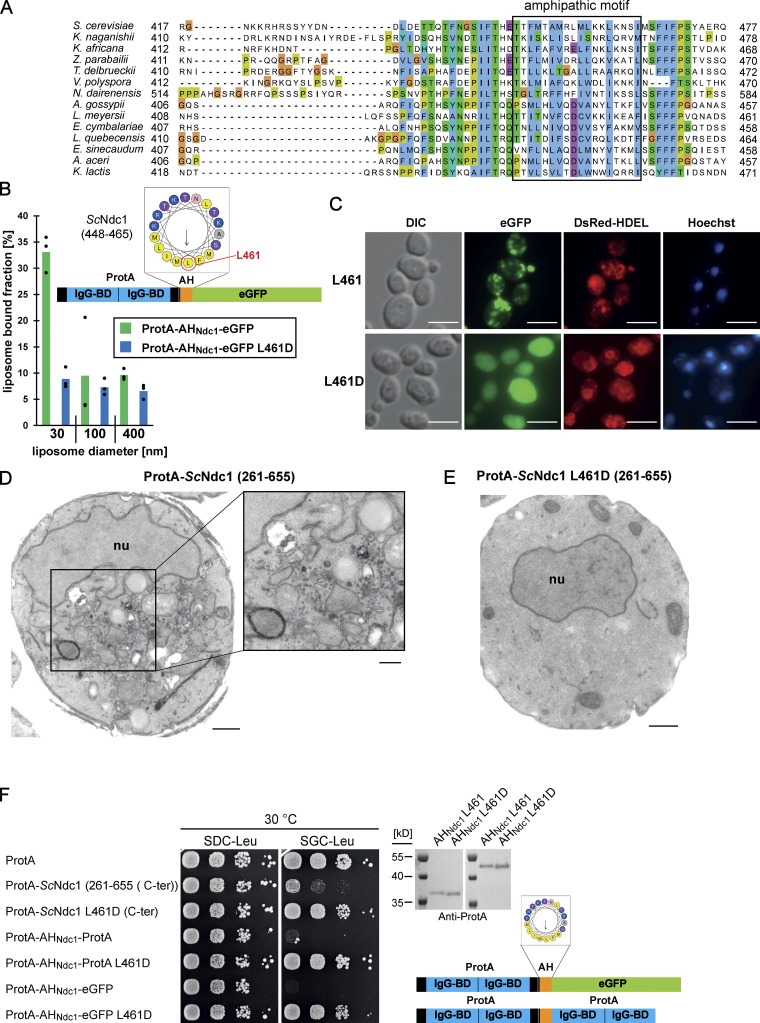
Figure 4.
The C-terminus of Ndc1 contains a conserved amphipathic sequence motif. (A) Multiple sequence alignment of a C-terminal region of Ndc1 homologs from different fungal species. The alignment was done using the ClustalW (Larkin et al., 2007) and displayed in Jalview (Waterhouse et al., 2009). The putative amphipathic regions in all sequences were detected using HELIQUEST (Gautier et al., 2008). (B) In vitro liposome binding assay with the isolated amphipathic helix (ProtA-AHNdc1-eGFP) and the corresponding L461D mutation using 30, 100, and 400 nm liposomes. Columns represent the mean of three independent experiments, individual data points are indicated. (C) Fluorescence microscopy of cells overexpressing the isolated amphipathic helix (ProtA-AHNdc1-eGFP) and the corresponding L461D mutation. DsRed-HDEL serves as ER marker. Chromatin is stained with Hoechst 33258. Bars, 5 µm. (D) Electron micrograph of a yeast cell overexpressing the C-terminal part of ScNdc1 (ProtA-ScNdc1 (261-655)) showing accumulation of cytoplasmic vesicles. Bar, 500 nm. In the right panel a higher magnification is shown (bar, 250 nm). (E) Electron micrograph of a yeast cell expressing ProtA-ScNdc1 (261-655) carrying the point mutation L461D. Bar, 500 nm. (F) Growth and expression were analyzed as in Fig. 3 E using yeast cells overexpressing the artificial model constructs ProtA-AHNdc1-ProtA and ProtA-AHNdc1-eGFP. The domain structures of both artificial constructs were schematically illustrated. Source data are available for this figure: SourceData F4.