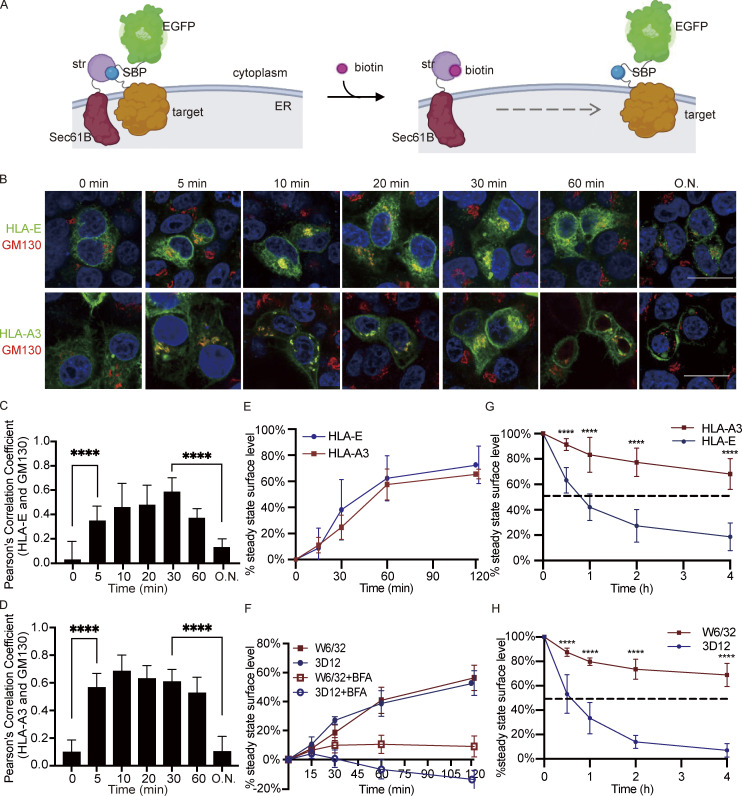
Figure 2.
Intracellular transport of HLA-E. (A) Schematic diagram of the RUSH system. The reporter complex contains the target protein (HLA-E or HLA-A3) with SBP and EGFP fused to the C terminus. The hook contains streptavidin (str) fused to the N terminus of the ER-resident protein Sec61B, which retains the reporter complex in the ER through the interaction between SBP and streptavidin. Biotin binds to streptavidin and enables the trafficking of the target protein out of ER. The figure was adapted from Boncompain et al. (2012) and created with BioRender.com. (B–D) HeLa cells were transiently cotransfected with Sec61B-streptavidin and HLA-E_SBP_EGFP or HLA-A3_SBP_EGFP. At different time points after biotin addition, cells were fixed, permeabilized, and stained with an antibody against the Golgi marker protein GM130, followed by detection with an Alexa568-conjugated secondary antibody. (B) Representative micrographs of two independent experiments. Scale bars = 20 μm. (C and D) Quantification of Golgi colocalization for HLA-E (C) or HLA-A3 (D). O.N., overnight. At each time point, PCC was calculated for 10–20 individual cells, and the data are shown as mean ± SD (error bars). (E) HEK 293T cells were transiently cotransfected with Sec61B-streptavidin and HLA-E_SBP_EGFP (blue circle) or HLA-A3_SBP_EGFP (red square). At different time points after biotin addition, cells were collected for flow cytometry analysis. The stable cell surface MFI after the biotin addition overnight was set to 100%, the MFI before biotin addition was set to 0%, and the other values were normalized accordingly. Data were collected for three biological runs and are shown as mean ± SD (error bars). (F) Surface HLA molecules on THP1-derived macrophages were stripped off by citric acid, and then cells were incubated in media with (hollow) or without (solid) BFA for different time points before flow cytometry analysis. The average cell surface MFI for time point zero was set to 0%, the average cell surface MFI without acid stripping was set to 100%, and the following values were normalized as its percentage. Data were collected for three biological runs and are shown as mean ± SD (error bars). (G and H) HeLa cells stably expressing HLA-A3 (red square) or HLA-E (blue circle; F) or THP1-derived macrophages (G) were incubated in media containing BFA for different time points, and surface expression of HLA molecules was assessed. The average cell surface MFI for time point zero was set to 100%, and the following values were normalized as their percentage. The dashed lines represent 50%. Data were collected for three biological runs and are shown as mean ± SD (error bars). Statistical analysis was performed using unpaired two-tailed Student’s t test with Welch’s correction. Asterisks show the statistical significance between indicated groups: ****, P < 0.0001.