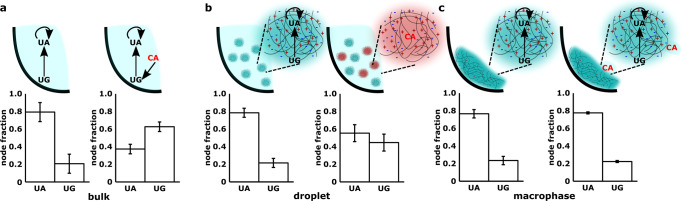
Fig. 4. Robustness of cross-catalytic RNA networks in coacervates.
Compositions are measured for a two-node cross-catalytic network in the absence or presence of a perturbing species under three different conditions. In this network, UA is self-catalyzed, as well as catalyzed by UG (via U → A link, a, top left). UG is also poorly self-catalyzed via U → G link (not shown). The perturbation is caused by adding a strong node (CA, shown in red) which catalyzes the synthesis of UG node (via C → G link, a, top right. a Bulk: Network is formed in the absence of any polymer and compositions are measured without perturbation (bottom left) as well as with perturbation (CA, bottom right). b Same as a but in coacervate droplets. Here, perturbing species (CA) is also encapsulated inside the coacervate droplets followed by mixing with droplets carrying the two-node network in 1:1 ratio, left without perturbation, right with perturbation. c Same as b but in coacervate macrophase. The perturbation is carried out by adding 0.5 μM solution of CA on top of macrophase, left without perturbation, right with perturbation. For each condition, samples were incubated for 30 min and then analyzed via polyacrylamide gel electrophoresis to measure the network compositions with or without perturbation. All the measurements are done in triplicates and mean WXYZ product formation is plotted as fraction w.r.t. each node along with standard deviation.