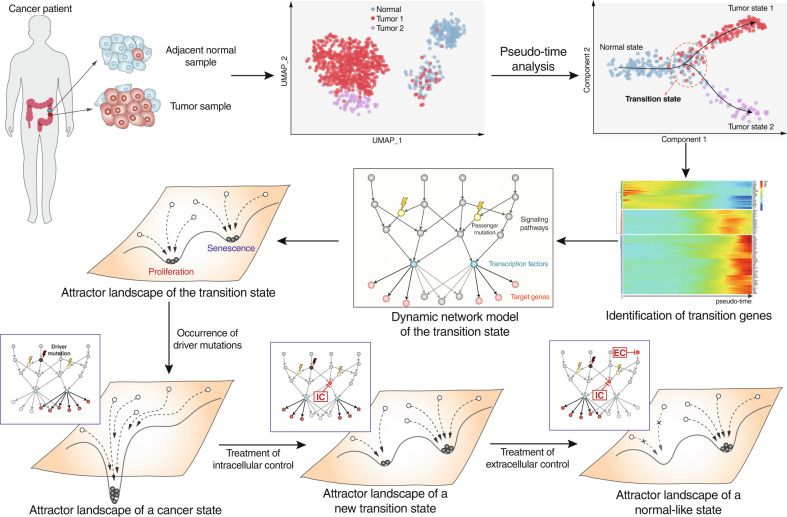
Fig. 7. Overview of ways to implement a control strategy for cancer reversion using single-cell genomic data and attractor landscape analysis.
Transcriptomes of single cells obtained from tumor and matched normal samples were clustered using dimension reduction techniques, and then cell identity was determined by measuring the expression levels of known cell marker genes. Cell populations, such as normal epithelial and tumor cells, can be ordered along tumorigenic trajectories using pseudotime analysis. The pseudotime information can also be used to construct a dynamic network model of the transition state through the identification of transition-related genes associated with tumorigenesis. Through a dynamic network model, the attractor landscape of a transition state can be identified. The attractor landscape of the cancer state can be determined by adding driver mutations to the network model. The dynamic network model and attractor landscape of cells in a cancer state can help to identify optimal IC and EC targets through the application of one or more control theories.