Figure 3. Semisupervised approach to extract coexpressed and flux‐dependent metabolic genes.
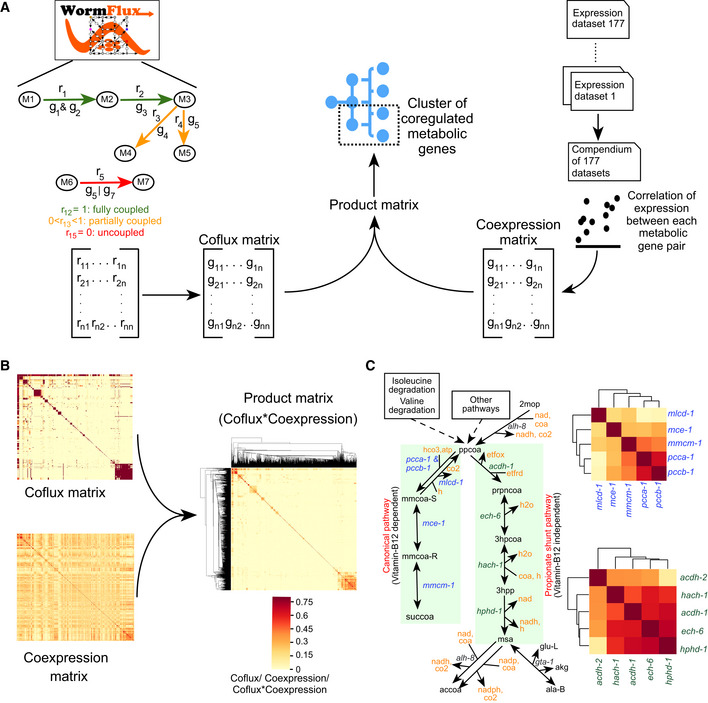
- Computational pipeline to extract tightly coregulated units in the metabolic network: Functional relationships are provided through theoretical flux associations (coflux) calculated using C. elegans metabolic network model iCEL1314, while expression correlations come from the compendium of 177 expression datasets. Hierarchical clustering on the product of coflux and coexpression matrix gives coexpressed metabolic pathways.
- Heatmaps showing coflux and coexpression of iCEL1314 genes and clustered heatmap showing added modularity to coexpression space by product of coexpression and coflux. Color legend is indicated.
- Distinct clusters denoted by clustered heatmap of genes in canonical and shunt pathways of propionate degradation were extracted using dynamic cut tree algorithm with stringent parameters (deepSplit = 2, minClusterSize = 3). Color legend as indicated in (B).
